The Surprising Speed of Compressed Data: A Roofline Story
Can a library designed for computing with compressed data ever hope to outperform highly optimized numerical engines like NumPy and Numexpr? The answer is complex, and it hinges on the "memory wall" — a phenomenon which occurs when system memory limitations start to drag on CPU. This post uses Roofline analysis to explore this very question, dissecting the performance of Blosc2 and revealing the surprising scenarios where it can gain a competitive edge.
TL;DR
Before we dive in, here's what we discovered:
For in-memory tasks, Blosc2's overhead can make it slower than Numexpr, especially on x86 CPUs.
This changes on Apple Silicon, where Blosc2's performance is much more competitive.
For on-disk tasks, Blosc2 consistently outperforms NumPy/Numexpr on both platforms.
The "memory wall" is real, and disk I/O is an even bigger one, which is where compression shines.
A Trip Down Memory Lane
Let's rewind to 2008. NumPy 1.0 was just a toddler, and the computing world was buzzing with the arrival of multi-core CPUs and their shiny new SIMD instructions. On the NumPy mailing list, a group of us were brainstorming how to harness this new power to make Python's number-crunching faster.
The idea seemed simple: trust newer compilers to use SIMD (and, possibly, data alignment) to perform operations on multiple data points at once. To test this, a simple benchmark was shared: multiply two large vectors element-wise. Developers from around the community ran the code and shared their results. What came back was a revelation.
For small arrays that fit snugly into the CPU's high-speed cache, SIMD was quite good at accelerating computations. But as soon as the arrays grew larger, the performance boost vanished. Some of us were already suspicious about the new "memory wall" that had been growing lately, seemingly due to the widening gap between CPU speeds and memory bandwidth. However, a conclusive answer (and solution) was still lacking.
But amidst the confusion, a curious anomaly emerged. One machine, belonging to NumPy legend Charles Harris, was consistently outperforming the rest—even those with faster processors. It made no sense. We checked our code, our compilers, everything. Yet, his machine remained inexplicably faster. The answer, when it finally came, wasn't in the software at all. Charles, a hardware wizard, had tinkered with his BIOS to overclock his RAM from 667 MHz to a whopping 800 MHz.
That was my lightbulb moment: for data-intensive tasks, raw CPU clock speed was not the limiting factor; memory bandwidth was what truly mattered.
This led me to a wild idea: what if we could make memory effectively faster? What if we could compress data in memory and decompress it on-the-fly, just in time for the CPU? This would slash the amount of data being moved, boosting our effective memory bandwidth. That idea became the seed for Blosc, a project I started in 2010 that has been my passion ever since. Now, 15 years later, it is time to revisit that idea and see how well it holds up in today's computing landscape.
Roofline Model: Understanding the Memory Wall
Not all computations are equally affected by the memory wall - in general performance can be either CPU-bound or memory-bound. To diagnose which resource is the limiting factor, the Roofline model provides an insightful analytical framework. This model plots computational performance against arithmetic intensity (i.e. floating-point operations per second versus memory accesses per second) to visually determine whether a task is constrained by CPU speed or memory bandwidth.
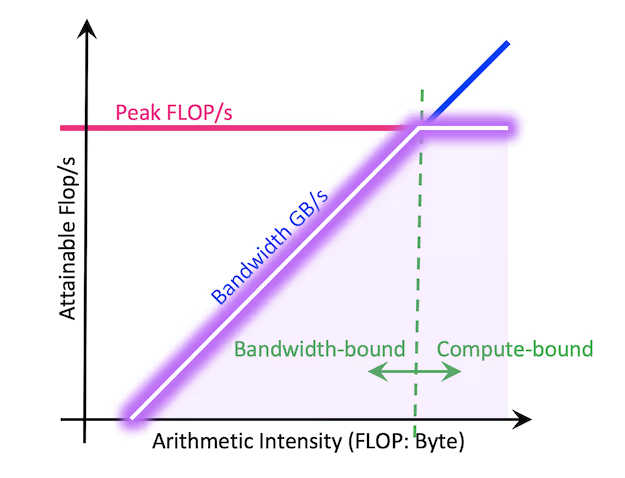
We will use Roofline plots to analyze Blosc2's performance, compared to that of NumPy and Numexpr. NumPy, with its highly optimized linear algebra backends, and Numexpr, with its efficient evaluation of element-wise expressions, together form a strong performance baseline for the full range of arithmetic intensities tested.
To highlight the role of memory bandwidth, we will conduct our benchmarks on an AMD Ryzen 7800X3D CPU at two different memory speeds: the standard 4800 MTS and an overclocked 6000 MTS. This allows us to directly observe how memory frequency impacts computational performance.
To cover a range of computational scenarios, our benchmarks include five operations with varying arithmetic intensities:
Very Low: A simple element-wise addition (a + b + c).
Low: A moderately complex element-wise expression (sqrt(a + 2 * b + (c / 2)) ^ 1.2).
Medium: A highly complex element-wise calculation involving trigonometric and exponential functions.
High: Matrix multiplication on small matrices (labeled matmul0).
Very High: Matrix multiplication on large matrices (labeled matmul1 and matmul2).
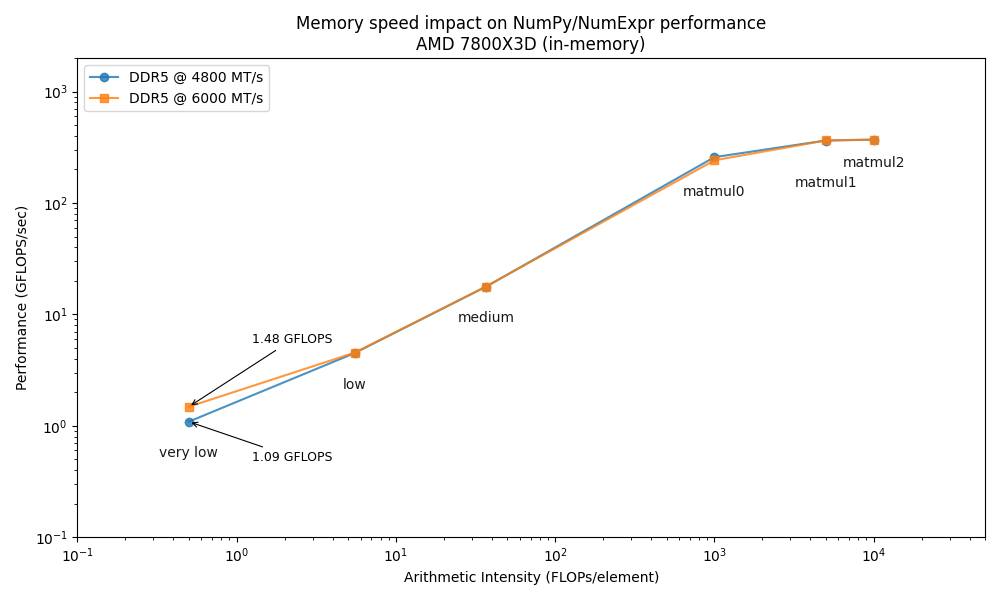
The Roofline plot confirms that increasing memory speed only benefits memory-bound operations (low arithmetic intensity), while CPU-bound tasks (high arithmetic intensity) are unaffected, as expected. Although this might suggest the "memory wall" is not a major obstacle, low-intensity operations like element-wise calculations, reductions, and selections are extremely common and often create performance bottlenecks. Therefore, optimizing for memory performance remains crucial.
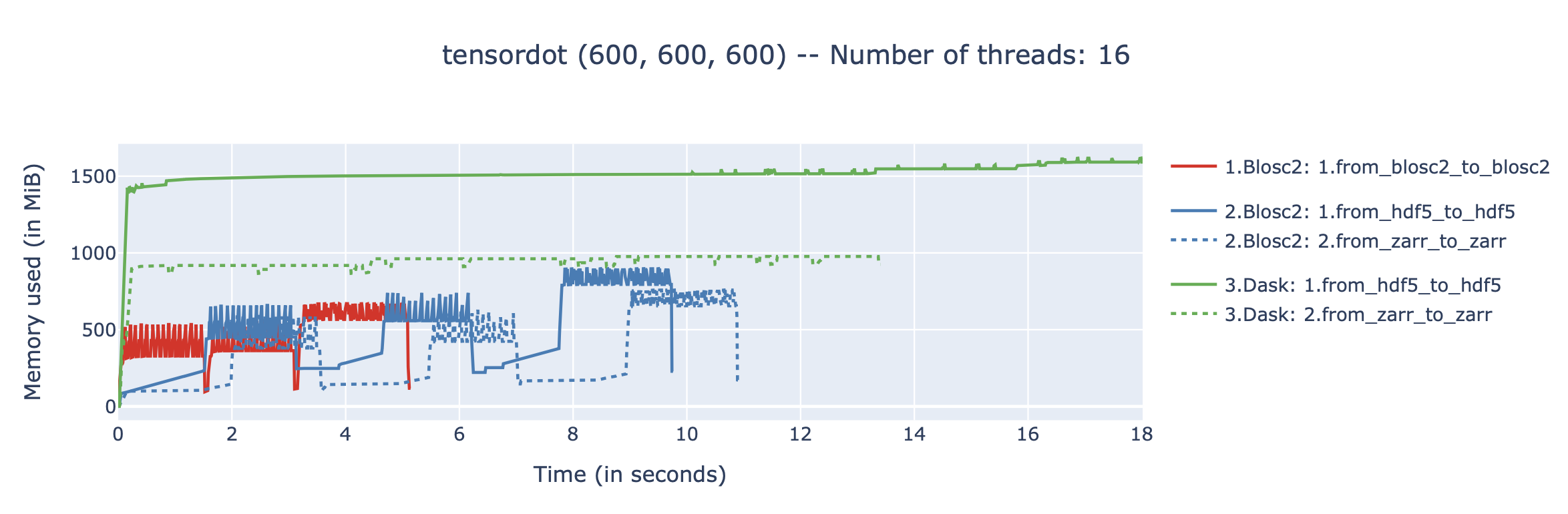
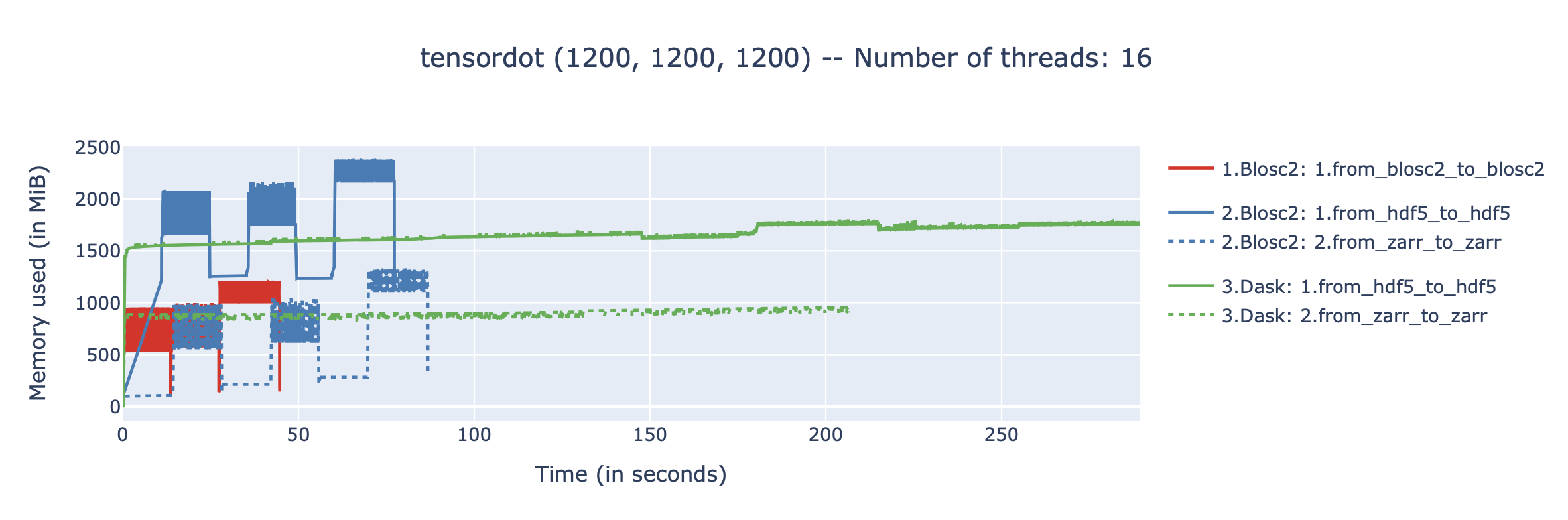
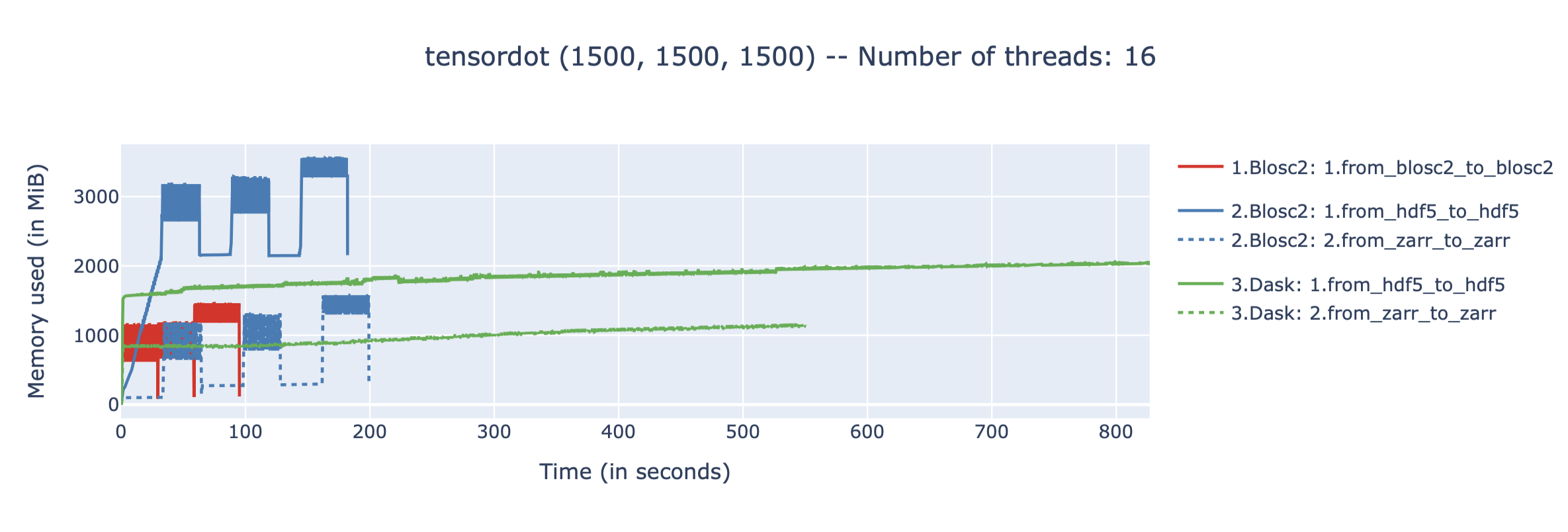
The In-Memory Surprise: Why Wasn't Compression Faster?
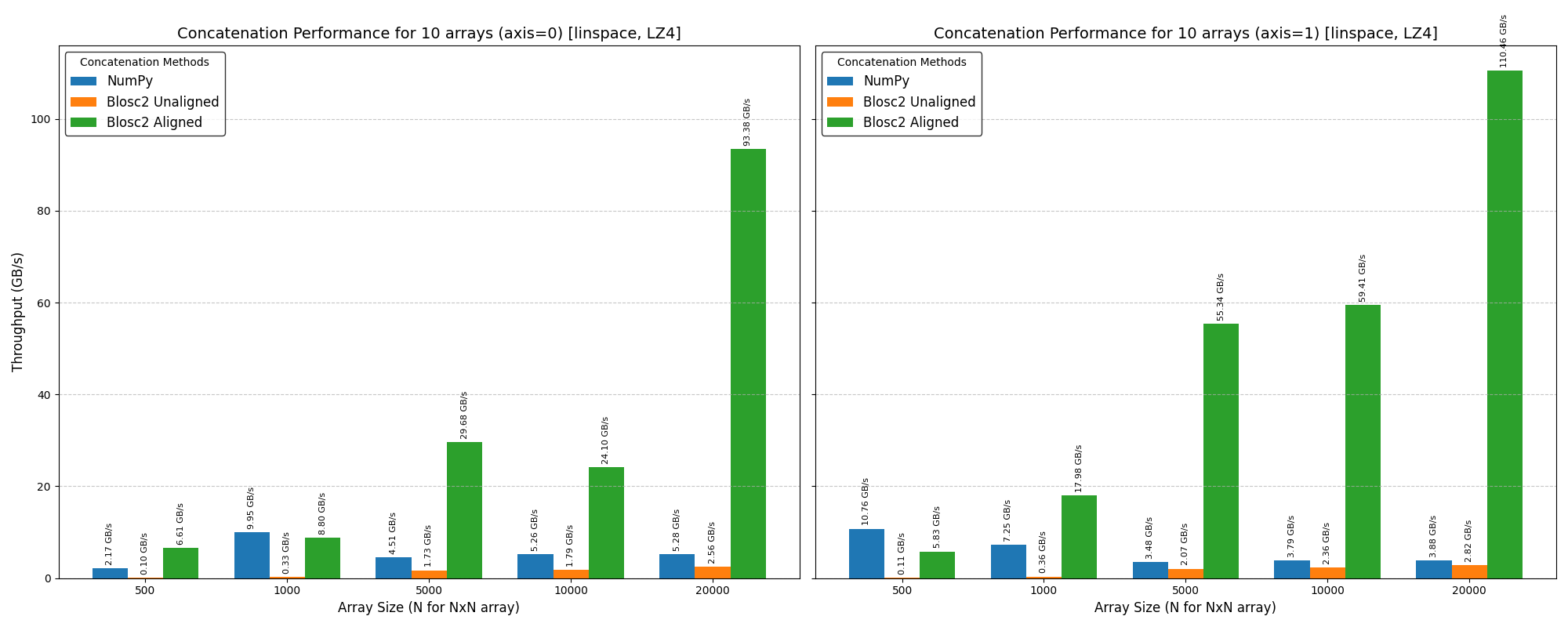
We benchmarked Blosc2 (both compressed and uncompressed) against NumPy and Numexpr. For this test, Blosc2 was configured with the LZ4 codec and shuffle filter, a setup known for its balance of speed and compression ratio. The benchmarks were executed on an AMD Ryzen 7800X3D CPU with memory speed set to 6000 MTS, ensuring optimal memory bandwidth for the tests.
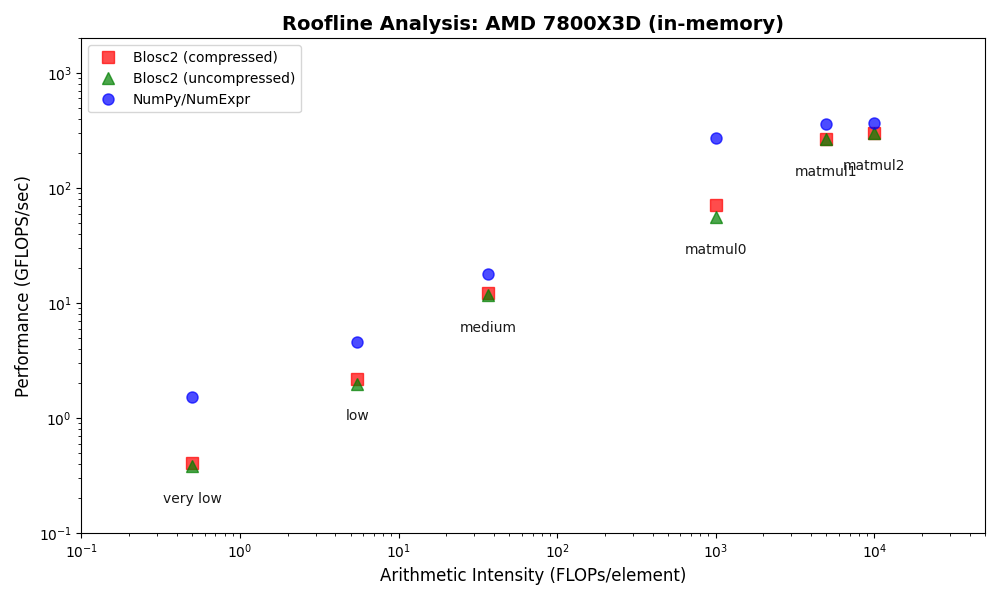
The analysis reveals a surprising outcome: for memory-bound operations, Blosc2 is up to five times slower than Numexpr. Although operating on compressed data provides a marginal improvement over uncompressed Blosc2, it is not enough to overcome this performance gap. This result is unexpected because Blosc2 leverages Numexpr internally, and the reduced memory bandwidth from compression should theoretically lead to better performance in these scenarios.
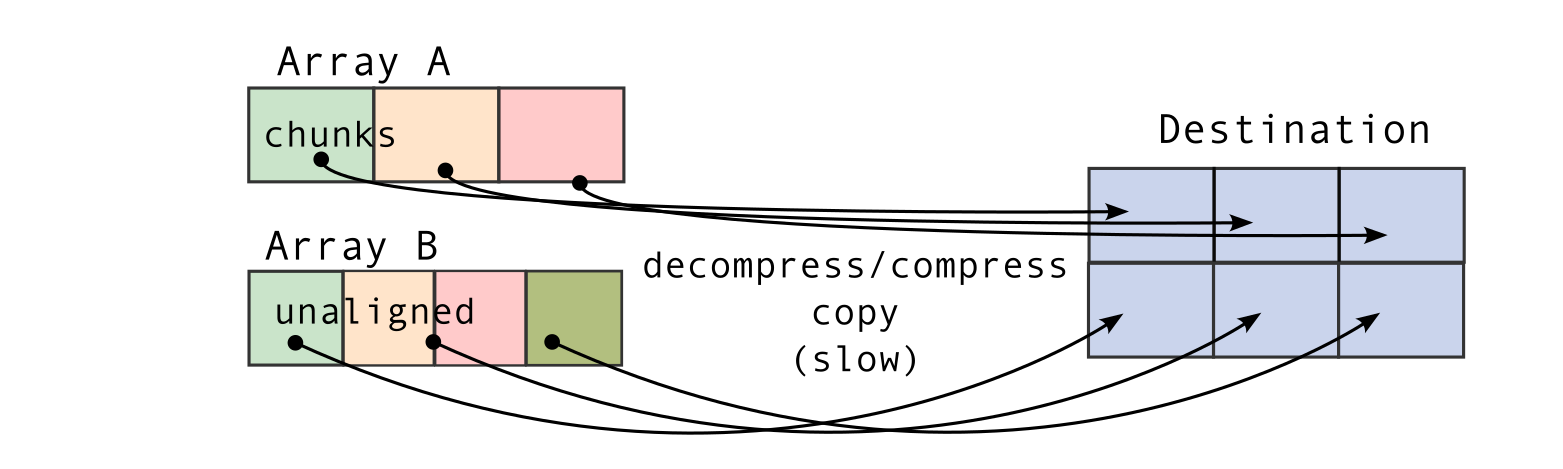
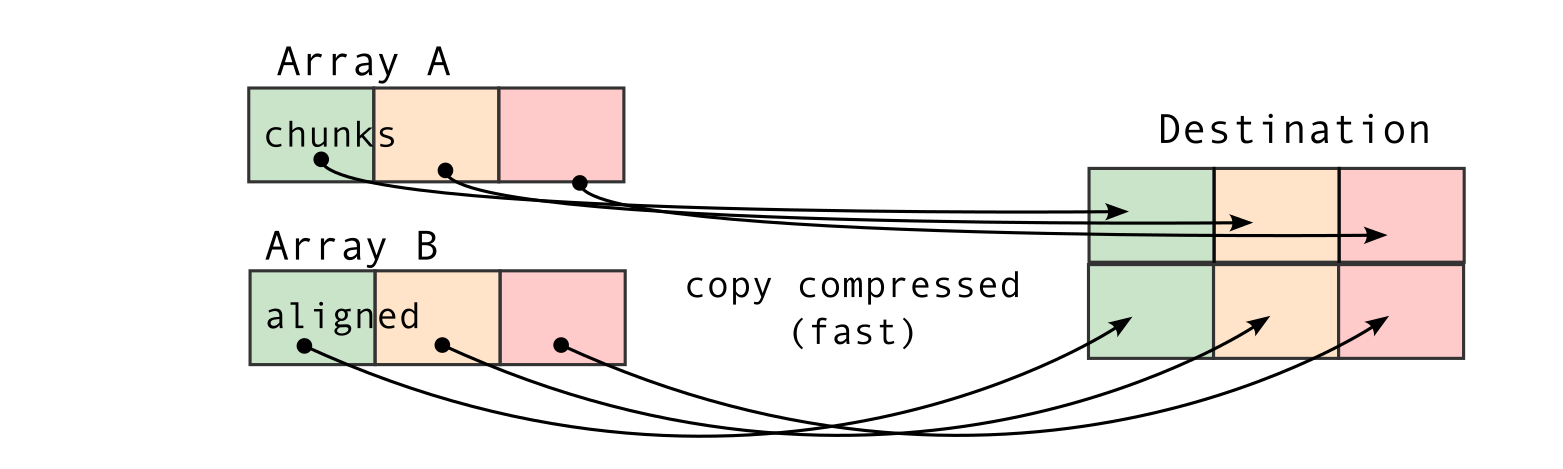
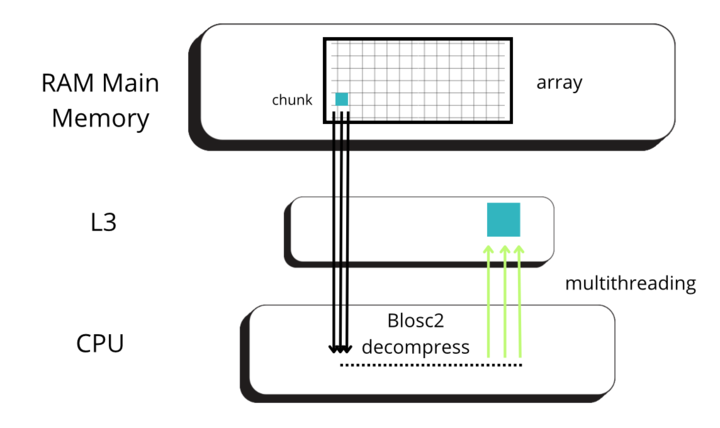
To understand this counter-intuitive result, we must examine Blosc2's core architecture. The key lies in its double partitioning scheme, which, while powerful, introduces an overhead that can negate the benefits of compression in memory-bound contexts.
Unpacking the Overhead: A Look Inside Blosc2's Architecture
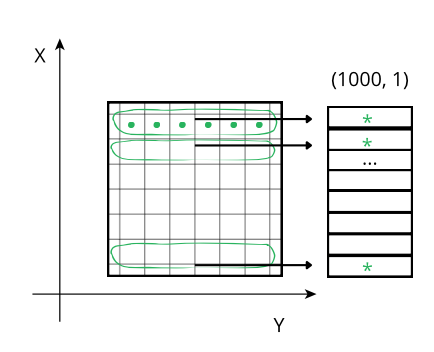
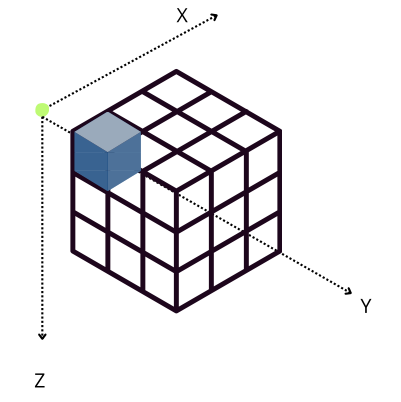
The performance characteristics of Blosc2 are rooted in its double partitioning architecture, which organizes data into chunks and blocks.
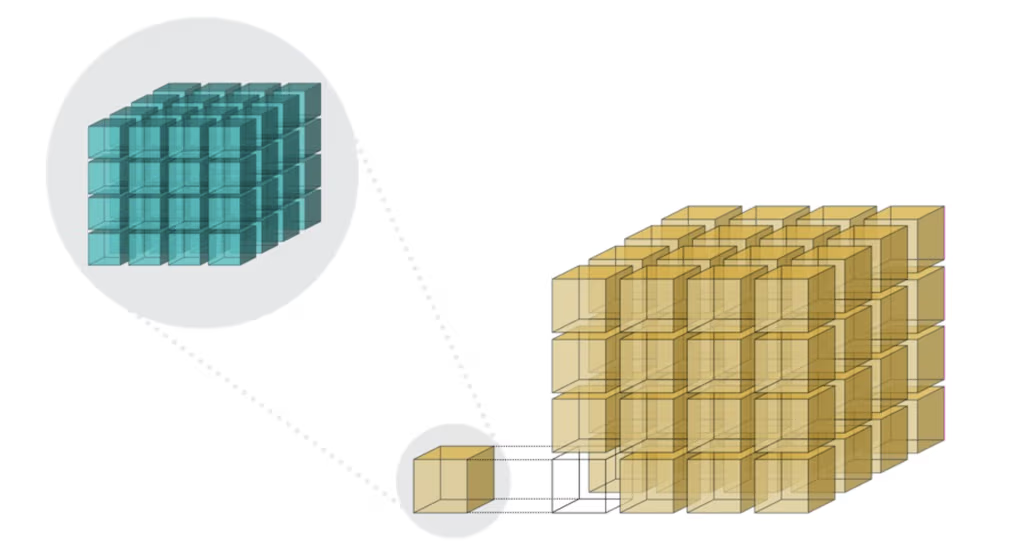
This design is crucial for both aligning with the CPU's memory hierarchy and enabling efficient multidimensional array representation (important for things like e.g. n-dimensional slicing). However, this structure introduces an inherent overhead from additional indexing logic. In memory-bound scenarios, this latency counteracts the performance gains from reduced memory traffic, explaining why Blosc2 does not surpass Numexpr.
Conversely, as arithmetic intensity increases, the computational demands begin to dominate the total execution time. In these CPU-bound regimes, the partitioning overhead is effectively amortized, allowing Blosc2 to close the performance gap and eventually match NumPy's performance in tasks like large matrix multiplications.
Modern ARM Architectures
CPU architecture is a rapidly evolving field. To investigate how these changes impact performance, we extended our analysis to the Apple Silicon M4 Pro, a modern ARM-based processor.
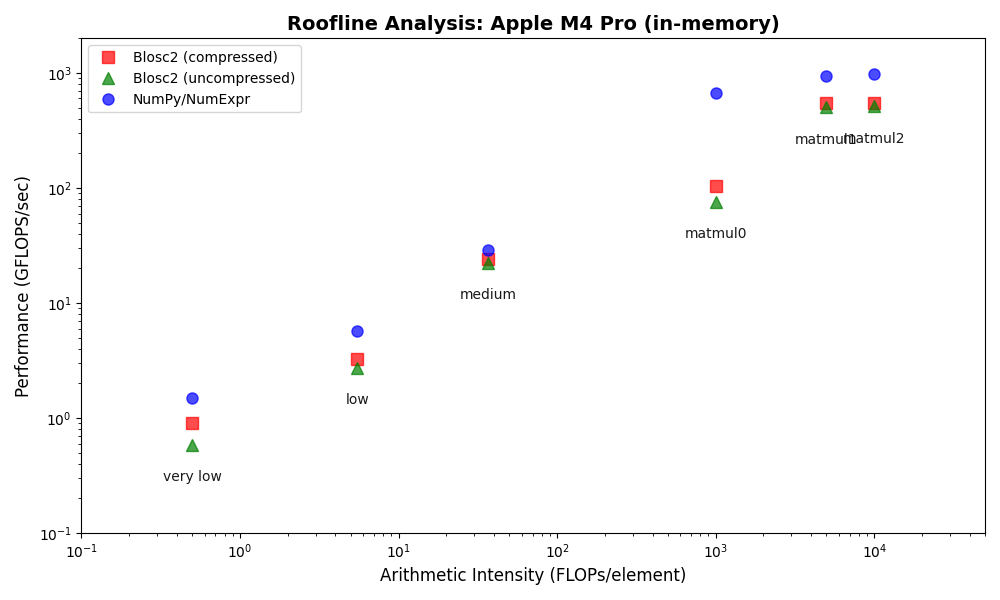
The results show that Blosc2 performs significantly better on this platform, narrowing the performance gap with NumPy/NumExpr, especially for operations on compressed data. While compute engines optimized for uncompressed data still hold an edge, these findings suggest that compression will play an increasingly important role in improving computational performance in the future.
However, while the in-memory results are revealing, they don't tell the whole story. Blosc2 was designed not just to fight the memory wall, but to conquer an even greater bottleneck: disk I/O. Although compression has the benefit of fitting more data into RAM when used in-memory (which is per se extremely interesting in these times, where RAM prices skyrocketed), its true power is unleashed when computations move off-motherboard. Now, let's shift the battlefield to the disk and see how Blosc2 performs in its native territory.
A Different Battlefield: Blosc2 Shines with On-Disk Data
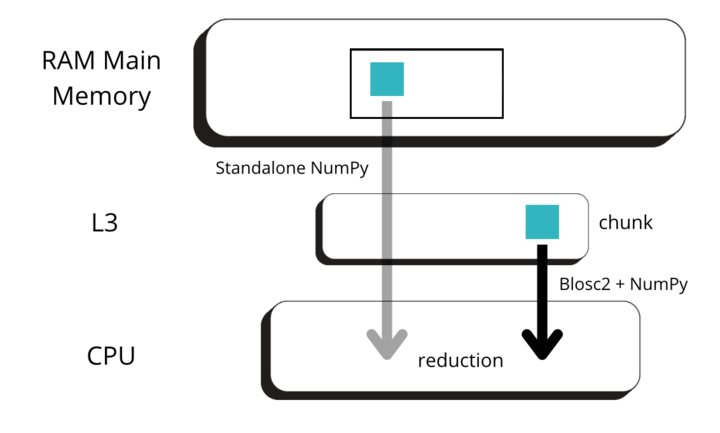
Blosc2's architecture extends its computational engine to operate seamlessly on data stored on disk, a significant advantage for large-scale analysis. This is particularly relevant in scenarios where datasets exceed available memory, necessitating out-of-core processing, as commonly encountered in data science, machine learning workflows or cloud computing environments.
Our on-disk benchmarks were designed to use datasets larger than the system's available memory to prevent filesystem caching from influencing the results. To establish a baseline, we implemented an out-of-core solution for NumPy/NumExpr, leveraging memory-mapped files. Here Blosc2 has a performance edge, particularly for memory-bound operations on compressed data, being able to send and receive data faster to disk than the memory-mapped NumPy arrays.
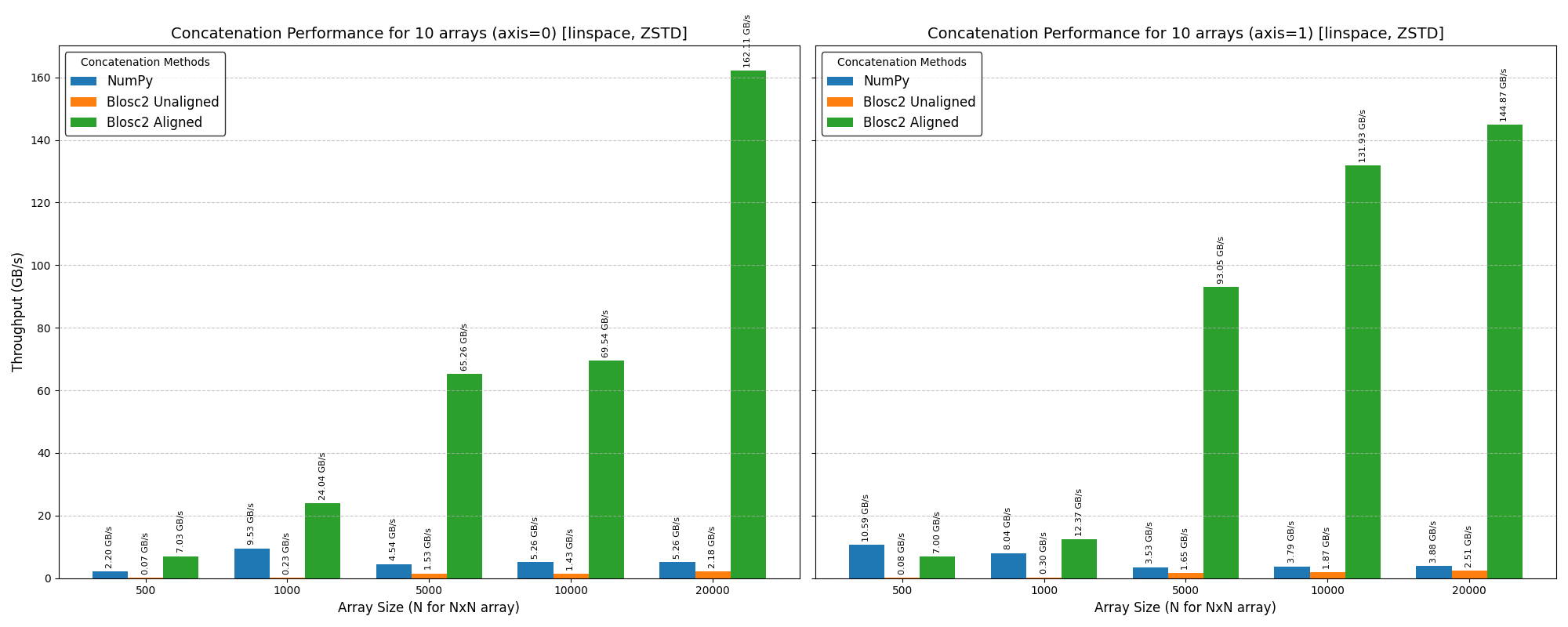
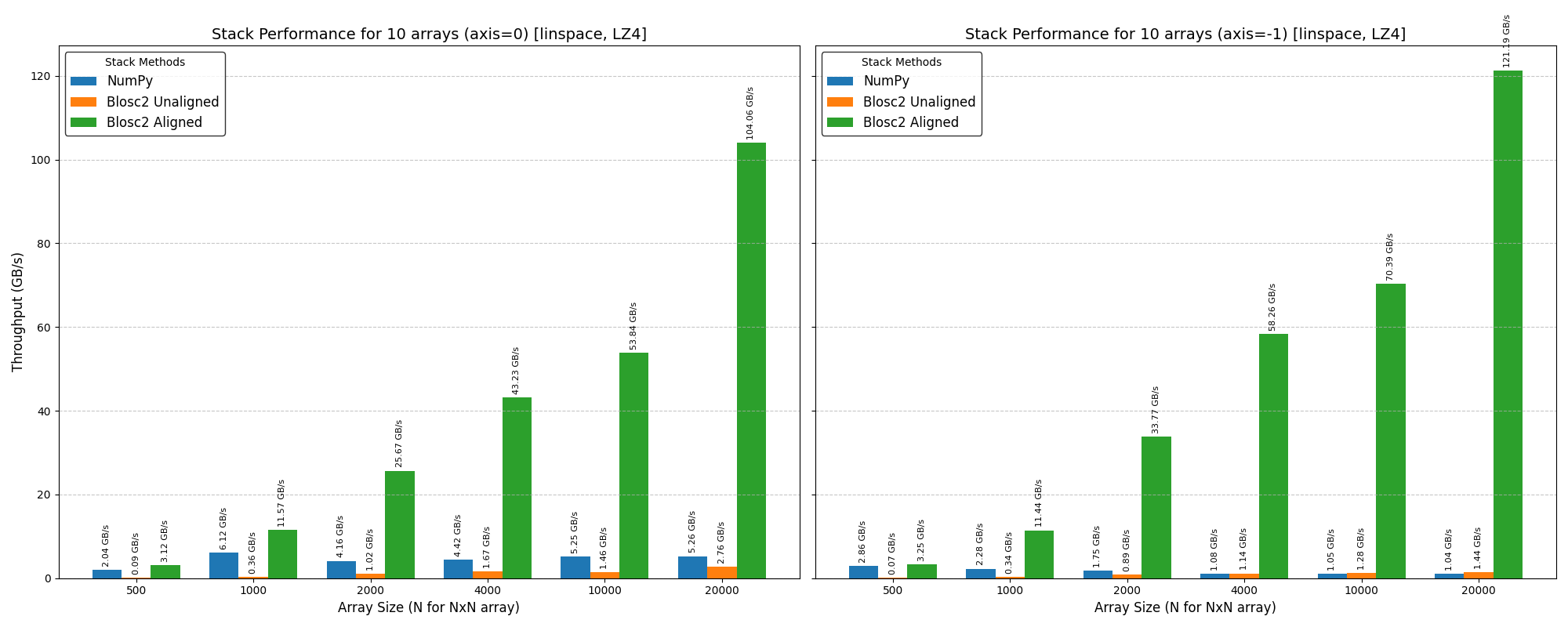
In this case, we've used high-performance NVMe SSDs (NVMe 4.0) to minimize the impact of disk speed on the results. We also switched to the ZSTD codec for Blosc2, as its superior compression ratio over LZ4 further minimizes data transfer to and from the disk.
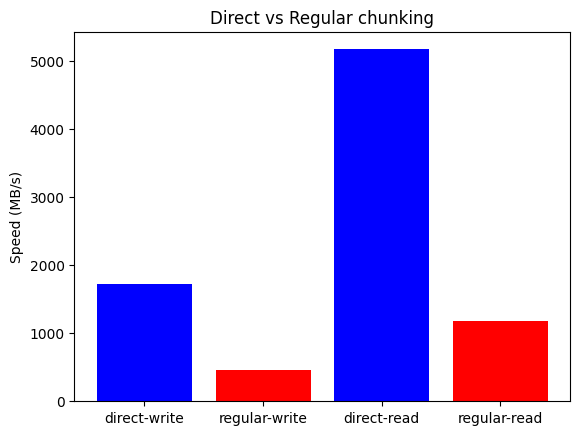
First, let's see the results for the AMD Ryzen 7800X3D system:

The plots above show that Blosc2 outperforms both NumPy and Numexpr for all low-to-medium intensity operations. This is because the high latency of disk I/O amortizes the overhead of Blosc2's double partitioning scheme. Furthermore, the reduced bandwidth required for compressed data gives Blosc2 an additional performance advantage in this scenario.
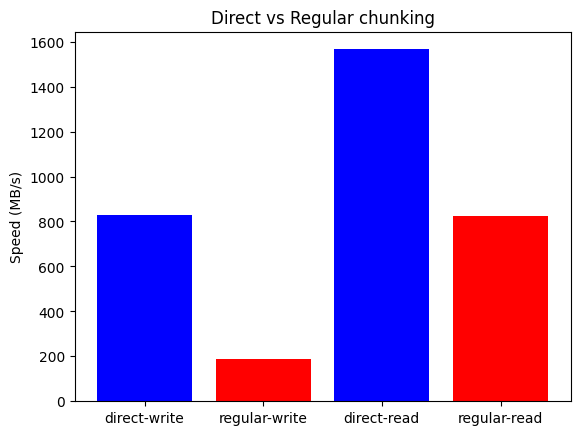
Now, let's see the results for the Apple Silicon M4 Pro system:
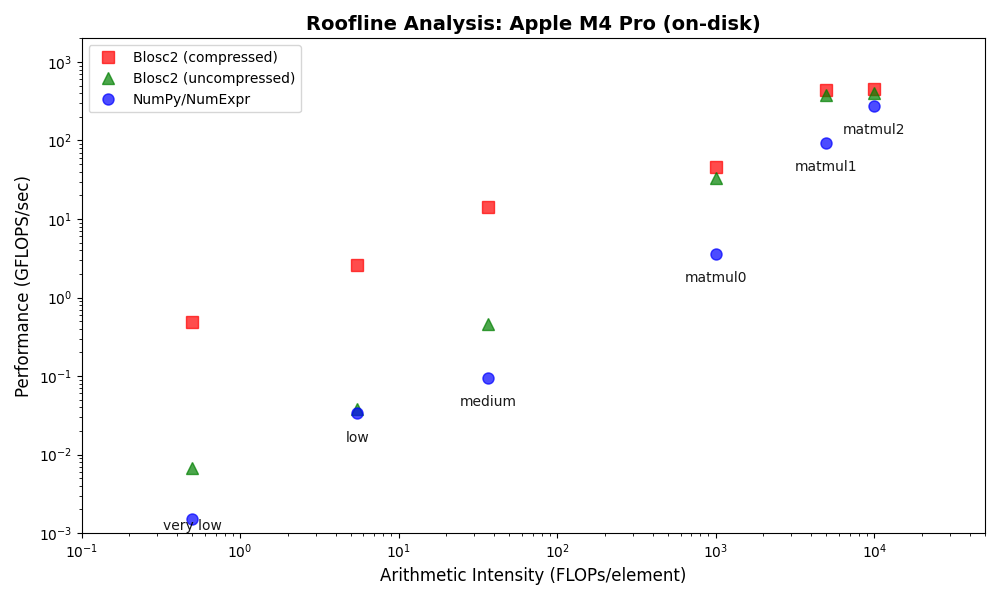
On the Apple Silicon M4 Pro system, Blosc2 again outperforms both NumPy and Numexpr for all on-disk operations, mirroring the results from the AMD system. However, the performance advantage is even more significant here, especially for memory-bound tasks. This is mainly because memory-mapped arrays are less efficient on Apple Silicon than on x86_64 systems, increasing the overhead for the NumPy/Numexpr baseline.
Roofline Plot: In-Memory vs On-Disk
To better understand the trade-offs between in-memory and on-disk processing with Blosc2, the following plot contrasts their performance characteristics for compressed data:
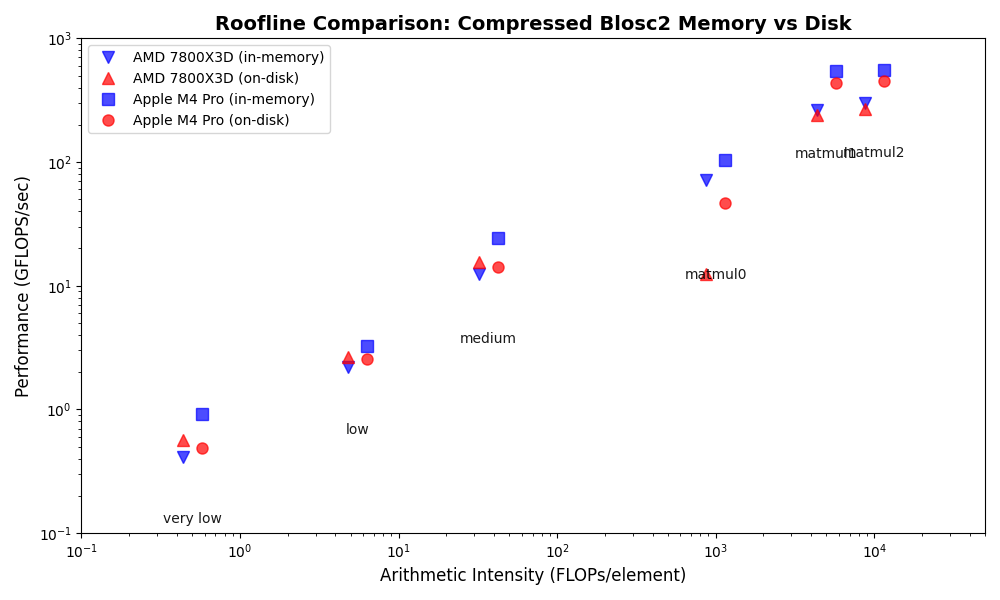
A notable finding for the AMD system is that Blosc2's on-disk operations are noticeably faster than its in-memory operations, especially for memory-bound tasks (low arithmetic intensity). This is likely due to two factors: first, the larger datasets used for on-disk tests allow Blosc2 to use more efficient internal partitions (chunks and blocks), and second, parallel data reads from disk further reduce bandwidth requirements.
In contrast, for CPU-bound tasks (high arithmetic intensity), on-disk performance is comparable to, albeit slightly slower than, in-memory performance. The analysis also reveals a specific weakness: small matrix multiplications (matmul0) are significantly slower on-disk, identifying a clear target for future optimization.
In contrast to the AMD system, the Apple Silicon M4 Pro shows that Blosc2's on-disk operations are slower than in-memory, a difference that is most significant for memory-bound tasks. This performance disparity suggests that current on-disk optimizations may favor x86_64 architectures over ARM.
As with the AMD platform, CPU-bound operations exhibit similar performance for both on-disk and in-memory contexts. The notable exception remains the small matrix multiplication (matmul0), which performs significantly worse on-disk. This recurring pattern pinpoints a clear opportunity for future optimization efforts.
Finally, and in addition to its on-disk performance, Blosc2 offers a significant cost advantage. With the recent rise in SSD prices, compressing data on disk becomes an economically attractive strategy, allowing you to store more data in less space and thereby reduce hardware expenses.
Reproducibility
All the benchmarks and plots presented in this blog post can be reproduced. You are invited to run the scripts on your own hardware to explore the performance characteristics of Blosc2 in different environments. In case you get interesting results, please consider sharing them with the community!
Conclusions
In this blog post, we explored the Roofline model to analyze the performance of Blosc2, NumPy, and Numexpr. We've confirmed that memory-bound operations are significantly affected by the "memory wall", making data compression of interest when maximizing performance. However, for in-memory operations, the overhead of Blosc2's double partitioning scheme can be a limiting factor, especially on x86_64 architectures. Encouragingly, this performance gap narrows considerably on modern ARM platforms like Apple Silicon, suggesting a promising future.
The situation changes dramatically for on-disk operations. Here, Blosc2 consistently outperforms NumPy and Numexpr, as the high latency of disk I/O (even if we used SSDs here) amortizes its internal overhead. This makes Blosc2 a compelling choice for out-of-core computations, one of its primary use cases.
Overall, this analysis has provided valuable insights, highlighting the importance of the memory hierarchy. It has also exposed specific areas for improvement, such as the performance of small matrix multiplications. As Blosc2 continues to evolve, I am confident we can address these points and further enhance its performance, making it an even more powerful tool for numerical computations in Python.
Read more about ironArray SLU — the company behind Blosc2, Caterva2, Numexpr and other high-performance data processing libraries.
Compress Better, Compute Bigger!