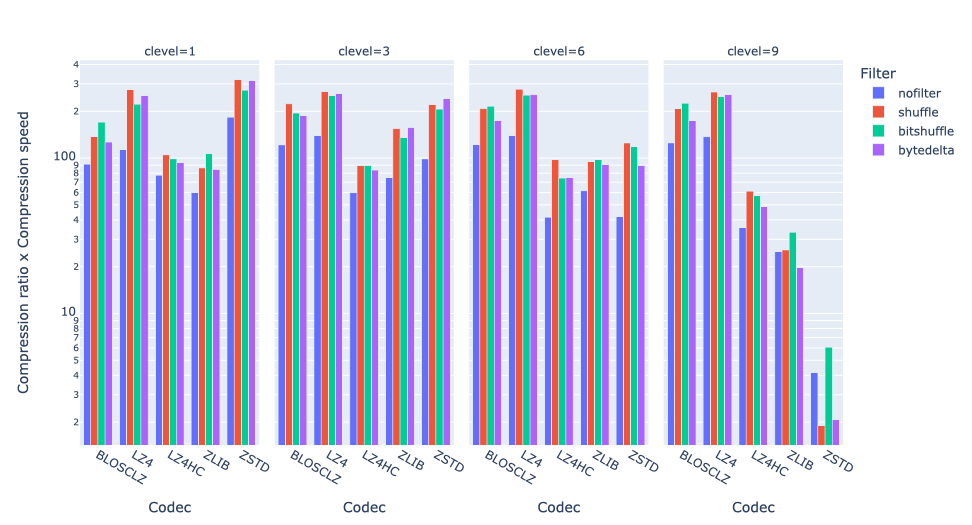
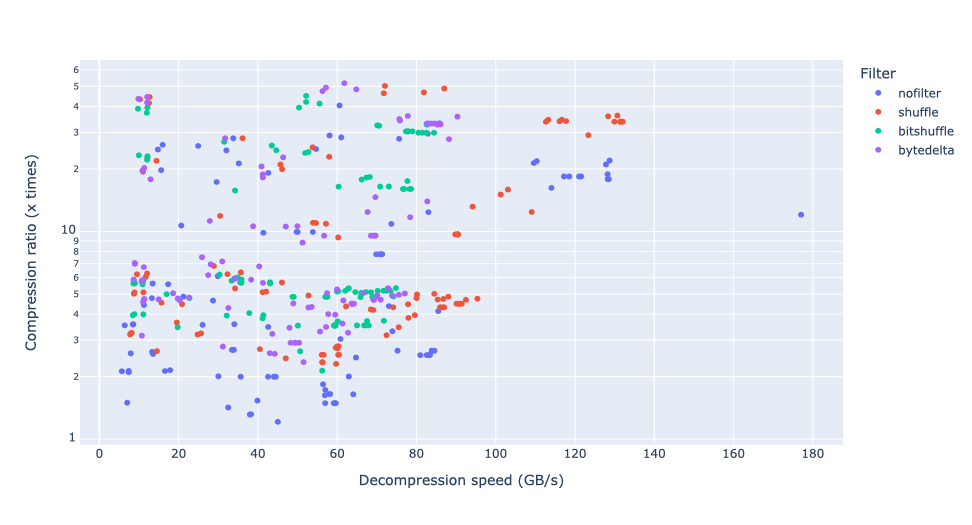
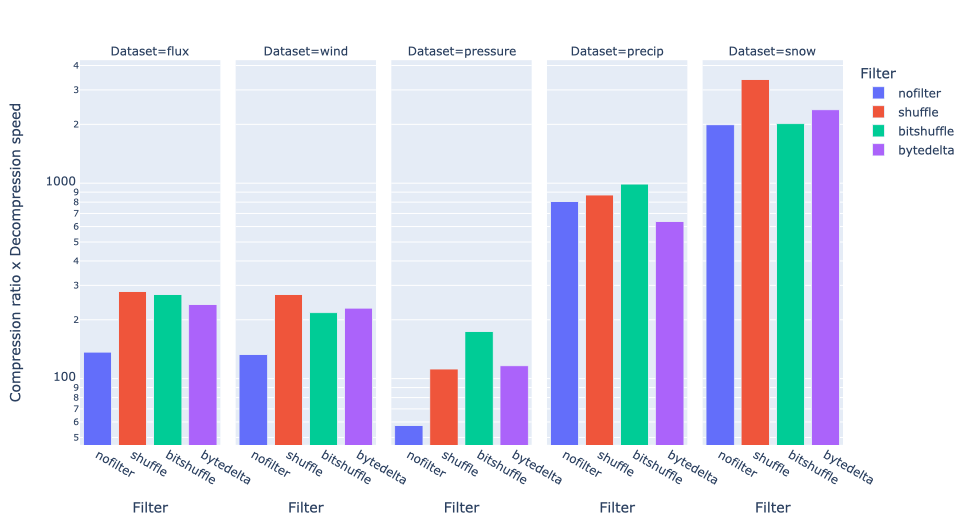
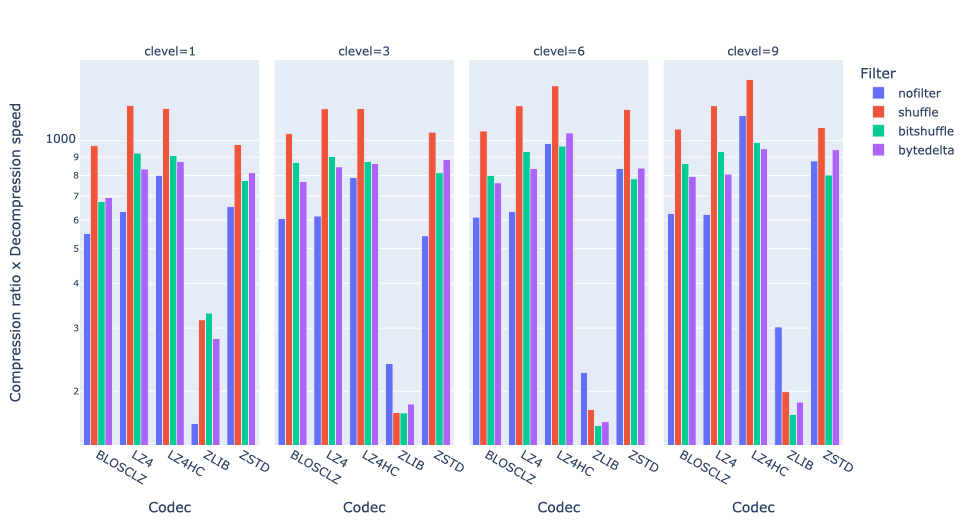
Exploring lossy compression with Blosc2
In the realm of data compression, efficiency is key. Whether you're dealing with massive datasets or simply aiming to optimize storage space and transmission speeds, the choice of compression algorithm can make a significant difference. In this blog post, we'll delve into the world of lossy compression using Blosc2, exploring its capabilities, advantages, and potential applications.
Understanding lossy compression
Unlike lossless compression, where the original data can be perfectly reconstructed from the compressed version, lossy compression involves discarding some information to achieve higher compression ratios. While this inevitably results in a loss of fidelity, the trade-off is often justified by the significant reduction in storage size.
Lossy compression techniques are commonly employed in scenarios where minor degradation in quality is acceptable, such as multimedia applications (e.g., images, audio, and video) and scientific data analysis. By intelligently discarding less crucial information, lossy compression algorithms can achieve substantial compression ratios while maintaining perceptual quality within acceptable bounds.
Lossy codecs in Blosc2
In the context of Blosc2, lossy compression can be achieved either through a combination of traditional compression algorithms and filters that can selectively discard less critical data, or by using codecs specially meant for doing so.
Filters for truncating precision
Since its inception, Blosc2 has featured the TRUNC_PREC filter, which is meant to discard the least significant bits from floating-point values (be they float32 or float64). This filter operates by zeroing out the designated bits slated for removal, resulting in enhanced compression. To see the impact on compression ratio and speed, an illustrative example here.
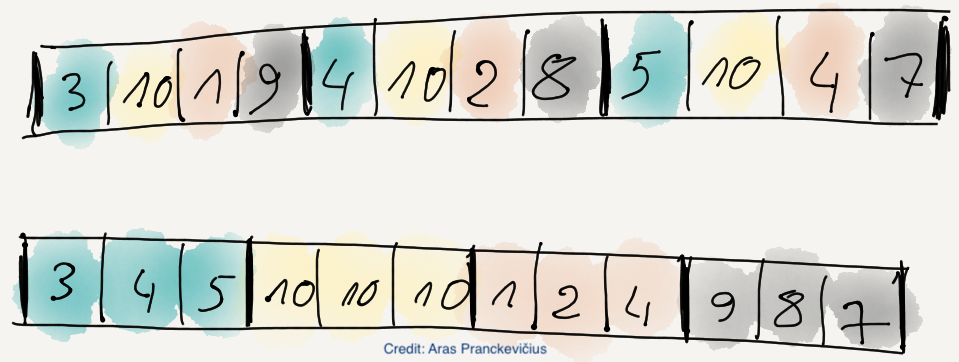
A particularly useful use case of the TRUNC_PREC filter is to truncate precision of float32/float64 types to either 8 or 16 bit; this is a quick and dirty way to ‘fake’ float8 or float16 types, which are very much used in AI nowadays, and contain storage needs.
In that vein, we recently implemented the INT_TRUNC filter, which does the same as TRUNC_PREC, but for integers (int8, int16, int32 and int64, and their unsigned counterparts). With both TRUNC_PREC and INT_TRUNC, you can specify an acceptable precision for most numerical data types.
Codecs for NDim datasets
Blosc2 has support for ZFP, another codec that is very useful for compressing multidimensional datasets. Although ZFP itself supports both lossless and lossy compression, Blosc2 makes use of its lossy capabilities only (the lossless ones are supposed to be already covered by other codecs in Blosc2). See this blog post for more info on the kind of lossy compression that can be achieved with ZFP.
Codecs for images
In addition, we recently included support for a couple of codecs that support the JPEG 2000 standard. One is OpenJ2HK, and the other is grok. Both have good, high quality JPEG 2000 implementations, but grok is a bit more advanced and has support for 16-bit gray images; we have blogged about it.
Experimental filters
Finally, you may want to experiment with some filters and codecs that were mainly designed to be a learning tool for people wanting to implement their own ones. Among them you can find:
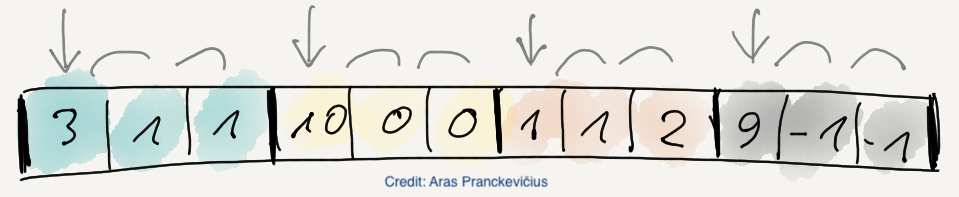
NDCELL: A filter that groups data in multidimensional cells, reordering them so that the codec can find better repetition patterns on a cell-by-cell basis.
NDMEAN: A multidimensional filter for lossy compression in multidimensional cells, replacing all elements in a cell by the mean of the cell. This allows for better compressions by the actual compression codec (e.g. NDLZ).
NDLZ: A compressor based on the Lempel-Ziv algorithm for 2-dim datasets. Although this is a lossless compressor, it is actually meant to be used in combination with the NDCELL and NDMEAN above, providing lossy compression for the latter case.
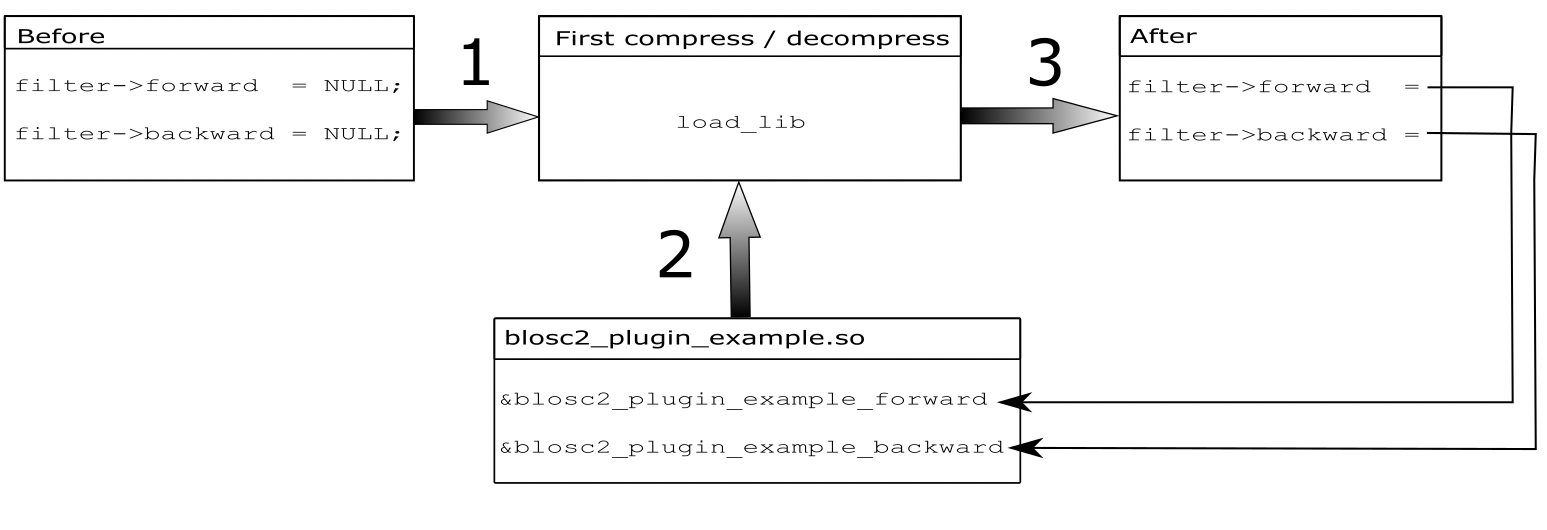
Again, the codecs in this section are not specially efficient, but can be used for learning about the compression pipeline in Blosc2. For more info on how to implement (and register) your own filters, see this blog post.
Applications and use cases
The versatility of Blosc2's lossy compression capabilities opens up a myriad of applications across different domains. In scientific computing, for example, where large volumes of data are generated and analyzed, lossy compression can significantly reduce storage requirements without significantly impacting the accuracy of results.
Similarly, in multimedia applications, such as image and video processing, lossy compression can help minimize bandwidth usage and storage costs while maintaining perceptual quality within acceptable limits.
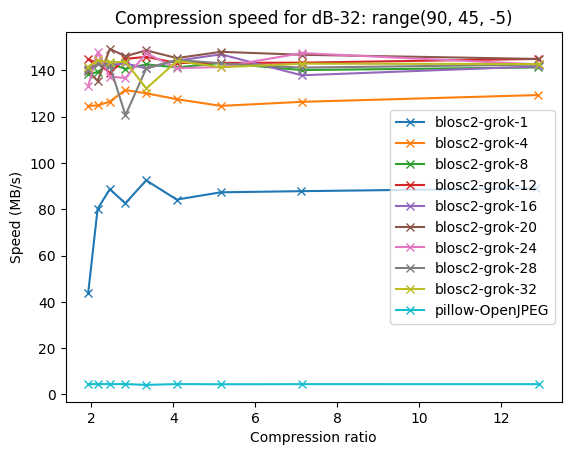
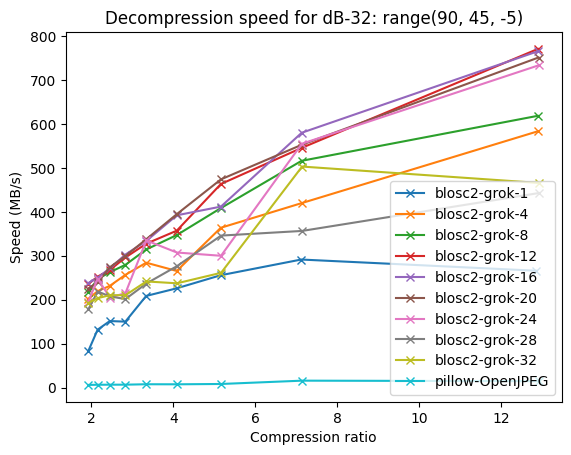
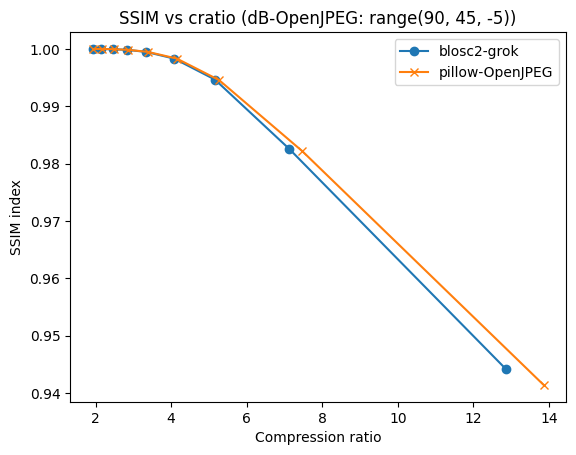
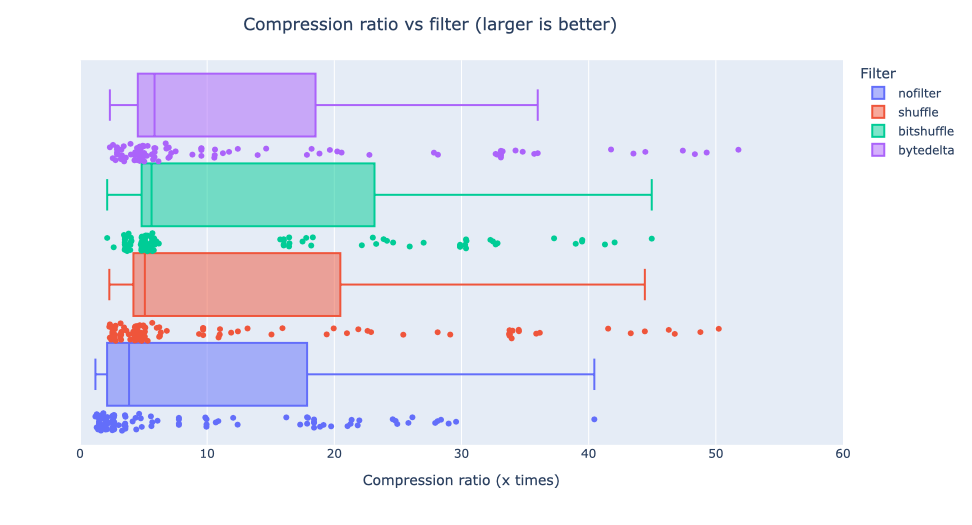
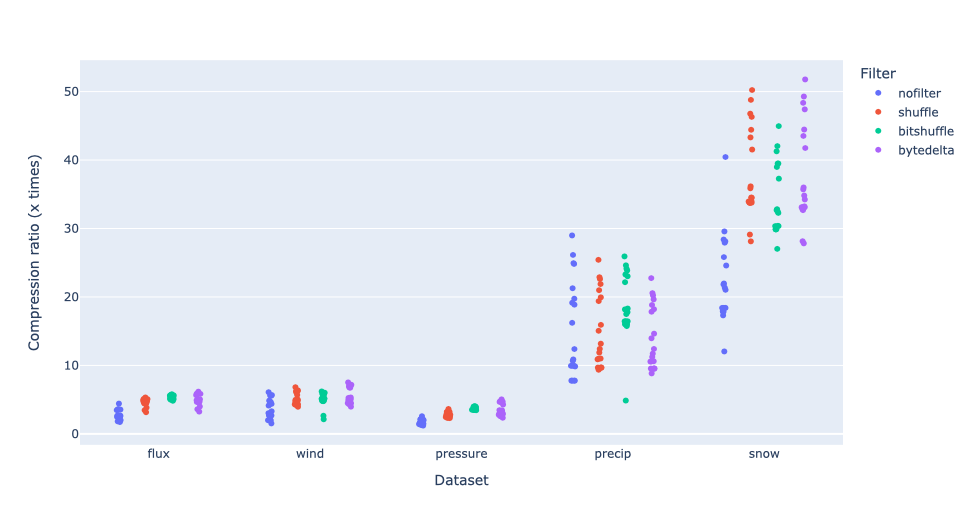
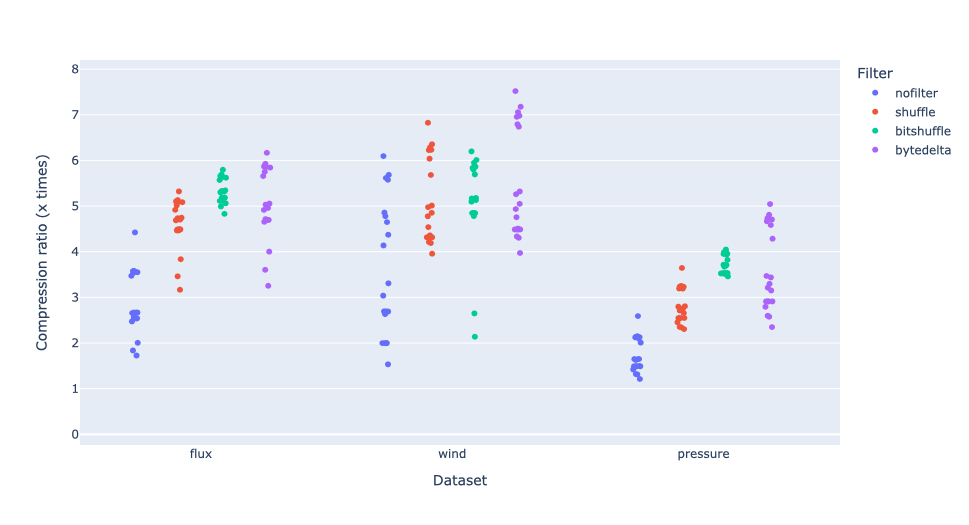
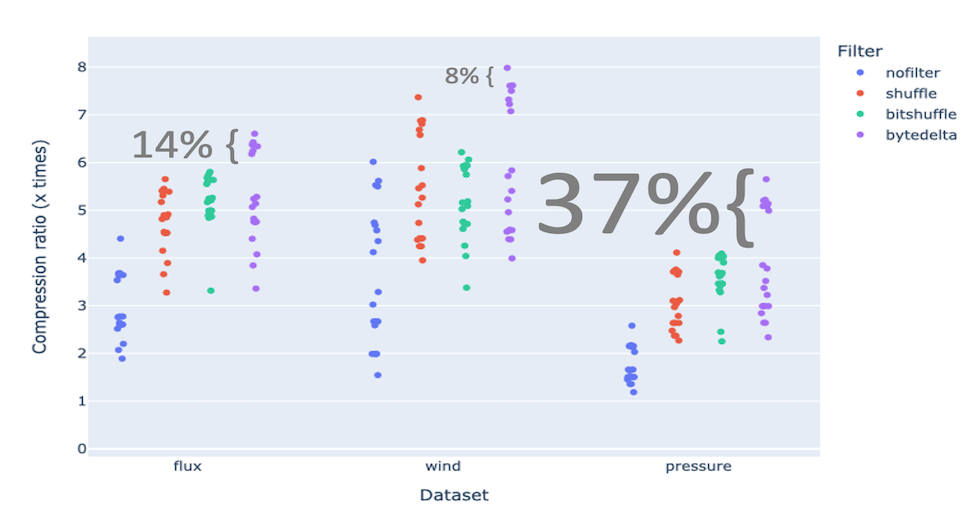
Compressing images with JPEG 2000 and with INT_TRUNC
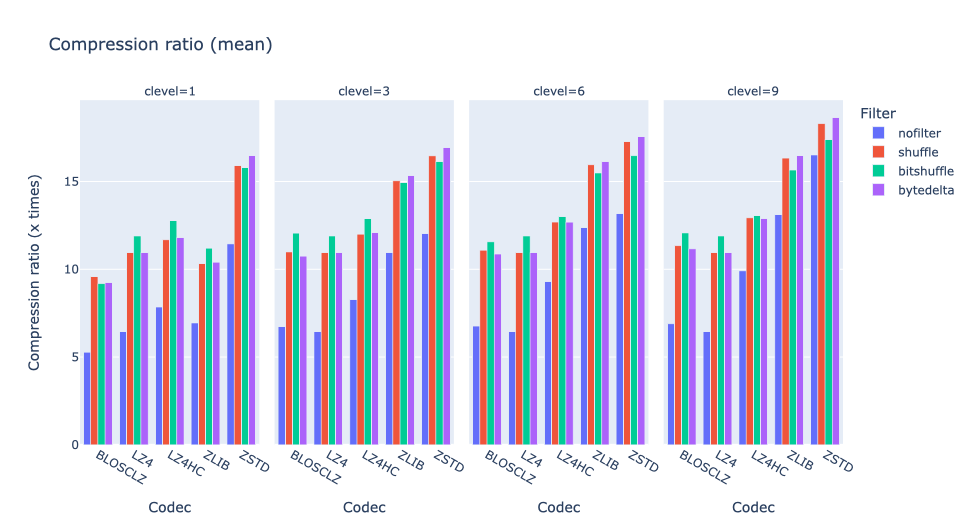
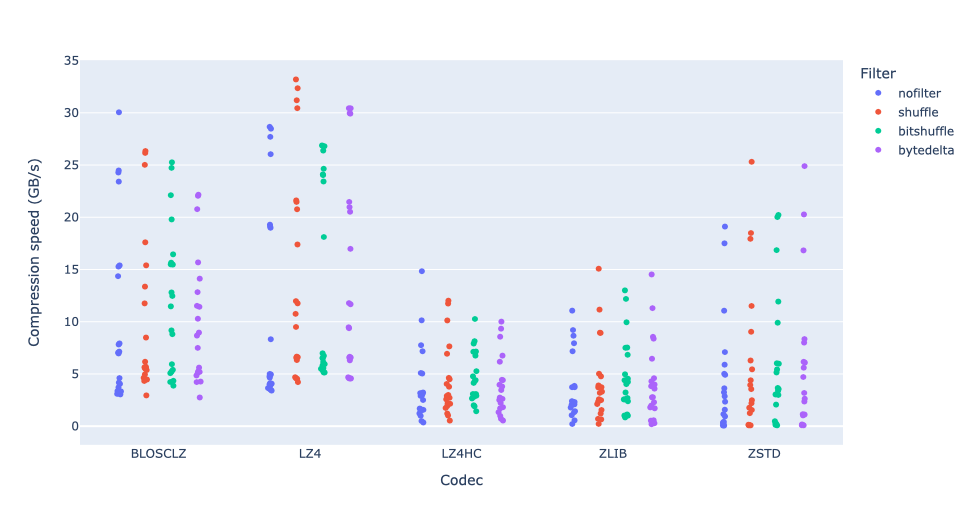
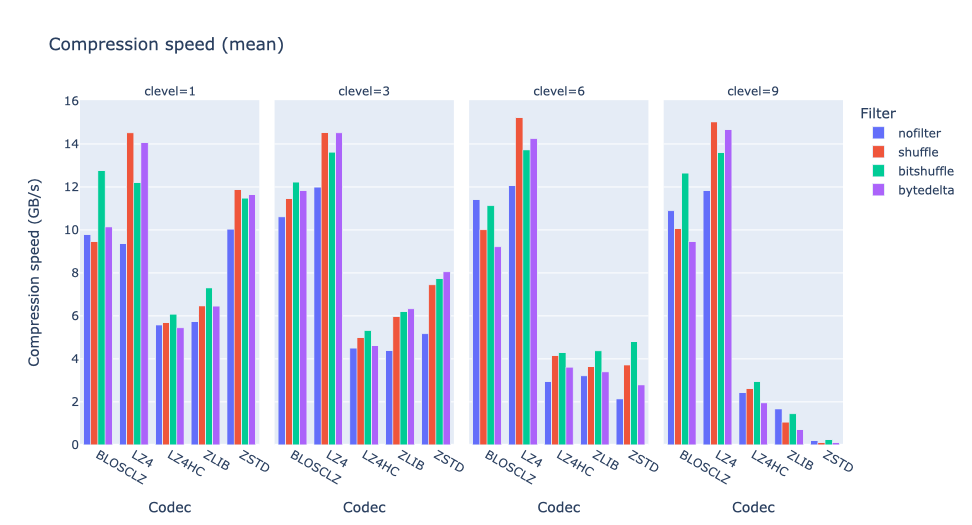
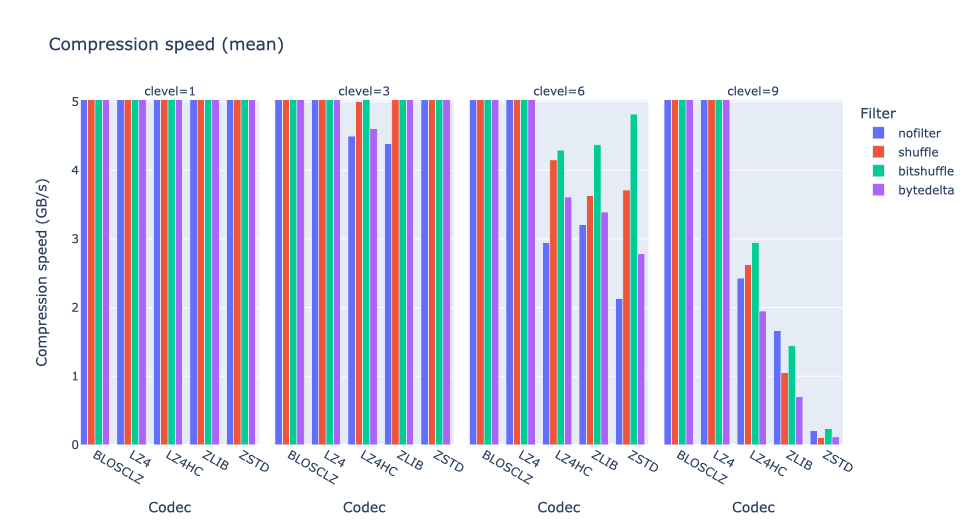
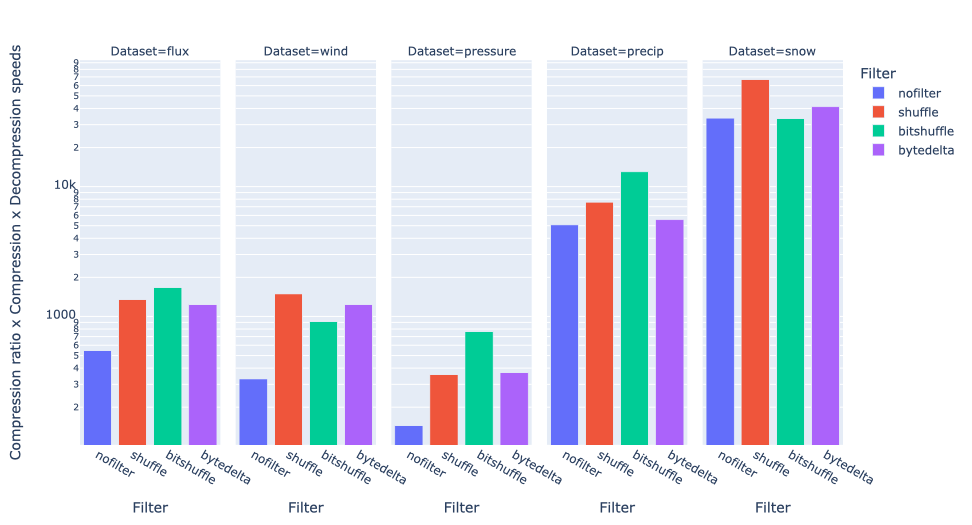
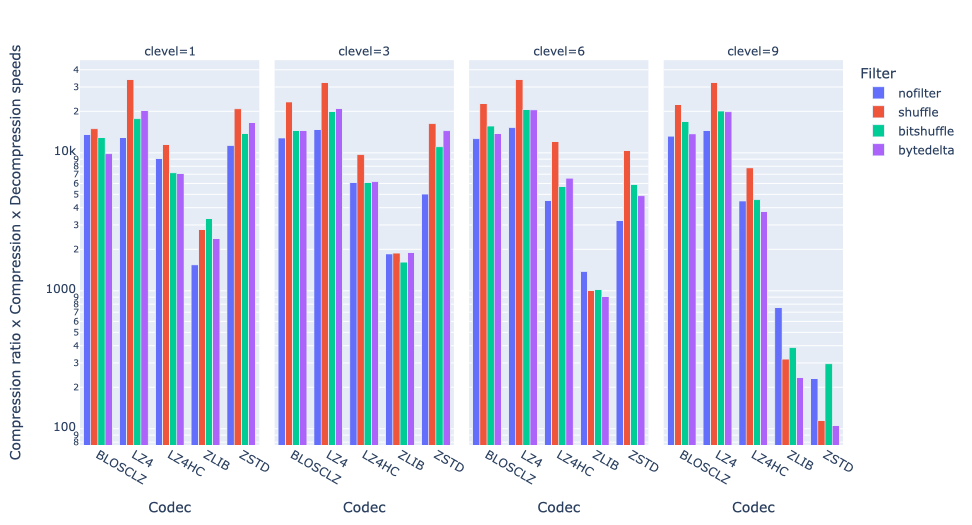
As an illustration, a recent study involved the compression of substantial volumes of 16-bit grayscale images sourced from different synchrotron facilities in Europe. While achieving efficient compression ratios necessitates the use of lossy compression techniques, it is essential to exercise caution to preserve key features for clear visual examination and accurate numerical analysis. Below, we provide an overview of how Blosc2 can employ various codecs and quality settings within filters to accomplish this task.
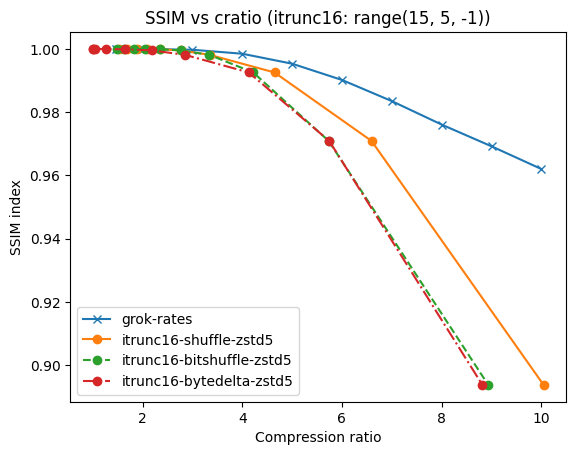
The SSIM index, derived from the Structural Similarity Measure, gauges the perceived quality of an image, with values closer to 1 indicating higher fidelity. You can appreciate the varying levels of fidelity achievable through the utilization of different filters and codecs.
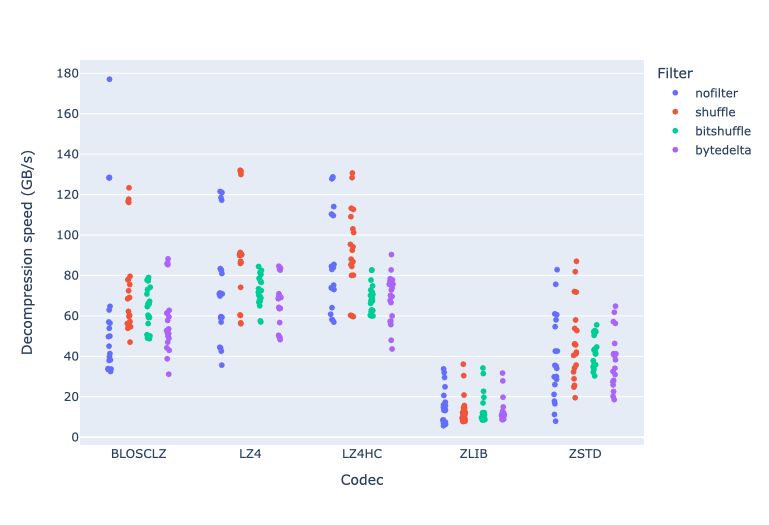
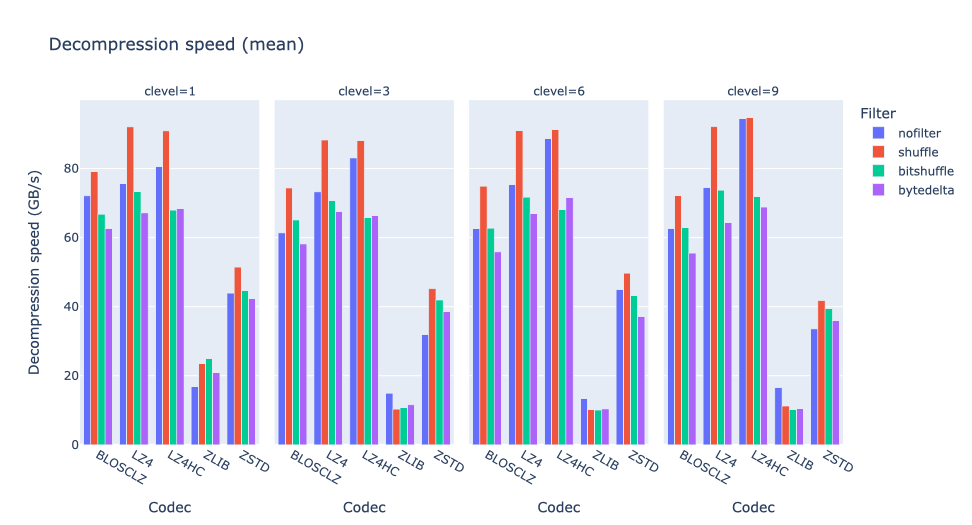
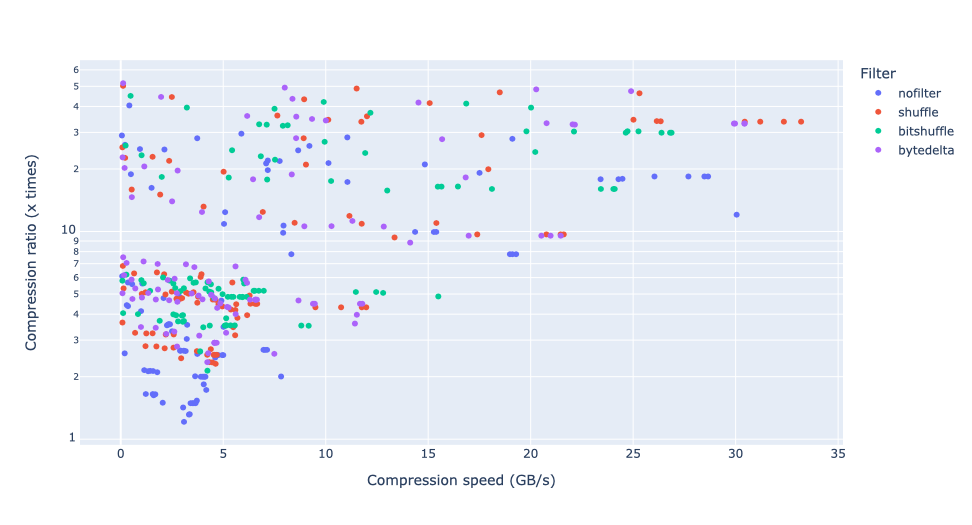
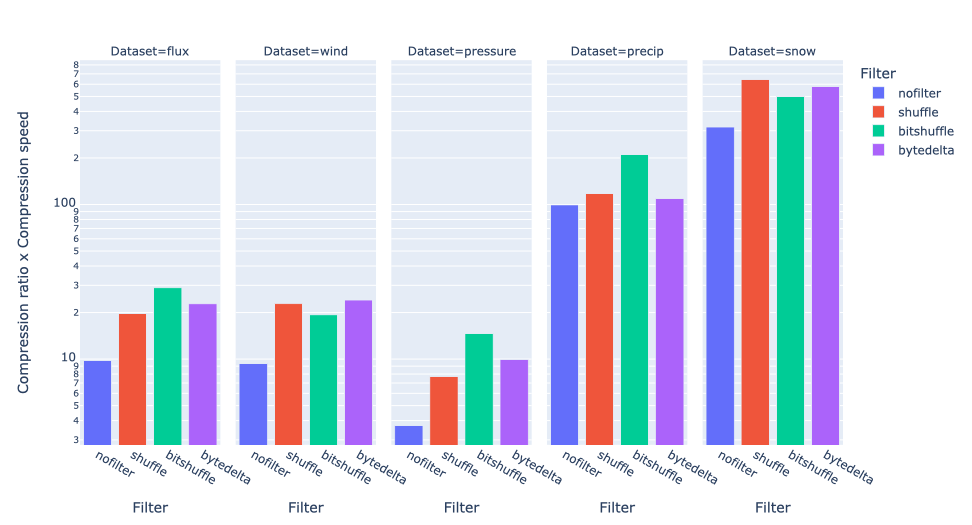
In terms of performance, each of these compression methods also showcases significantly varied speeds (tested on a MacBook Air with an M1 processor):
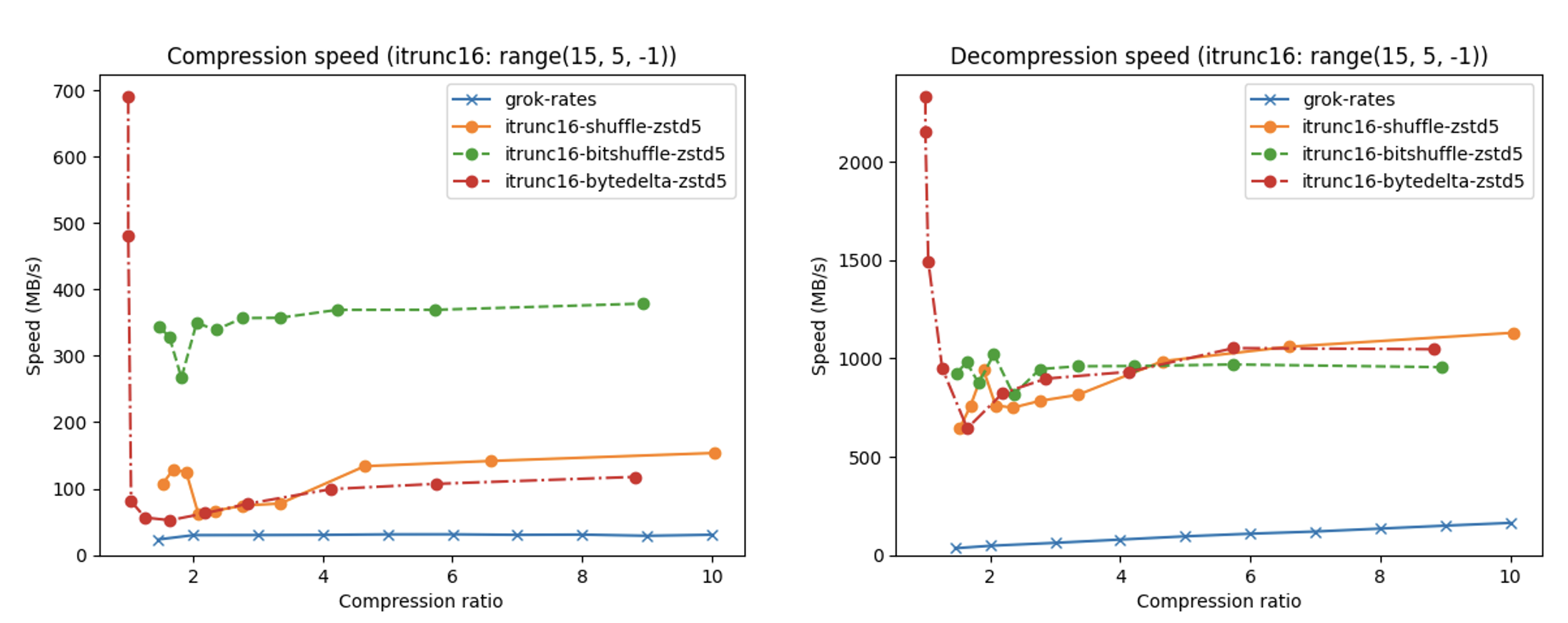
A pivotal benefit of Blosc2's strategy for lossy compression lies in its adaptability and configurability. This enables tailoring to unique needs and limitations, guaranteeing optimal performance across various scenarios.
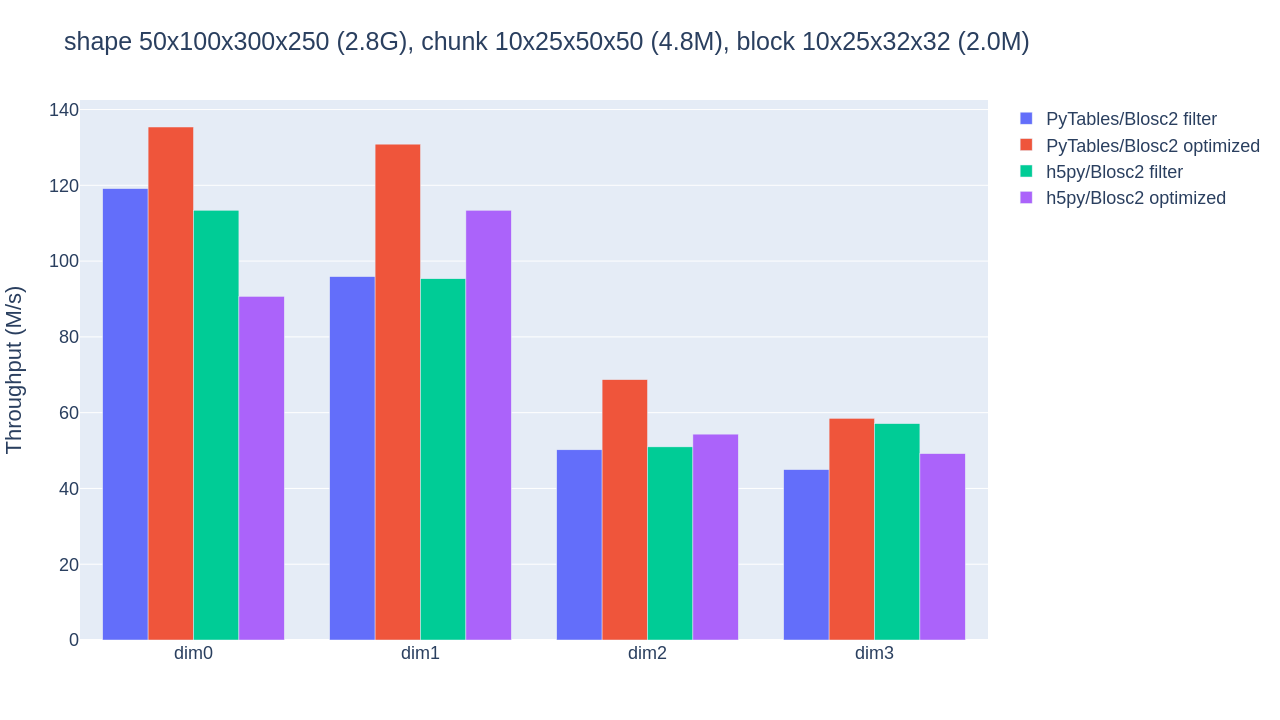
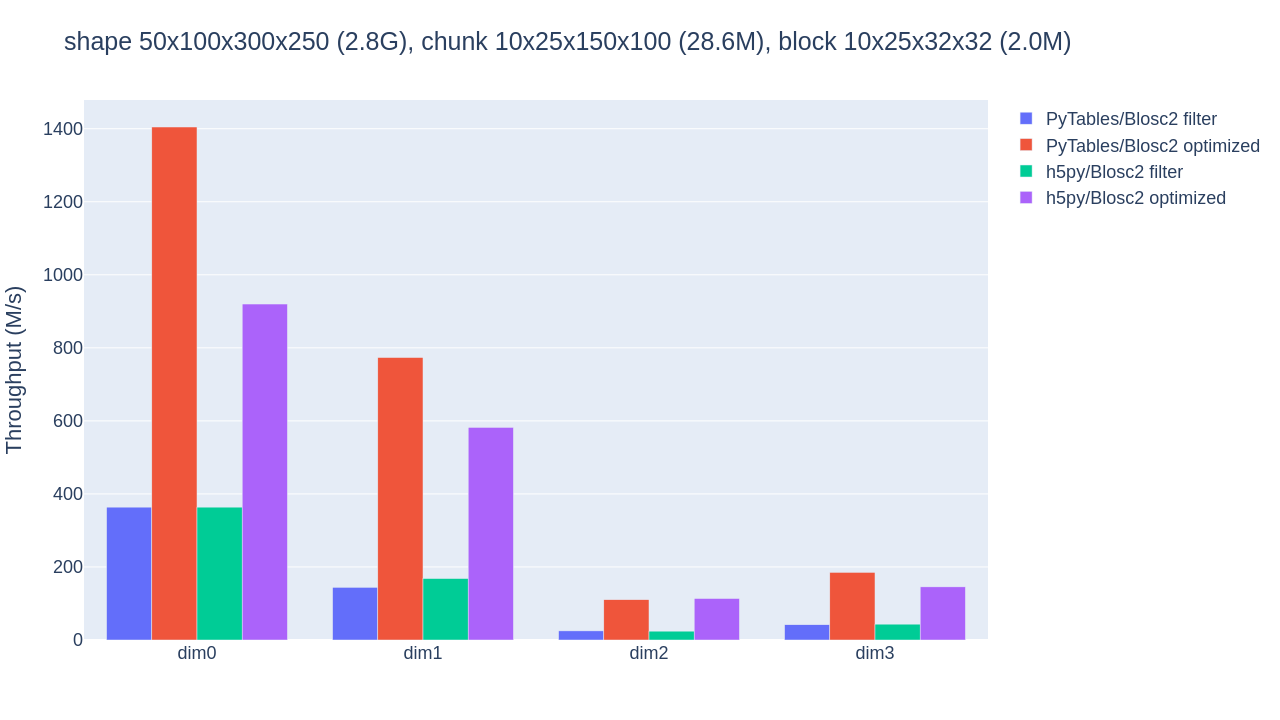
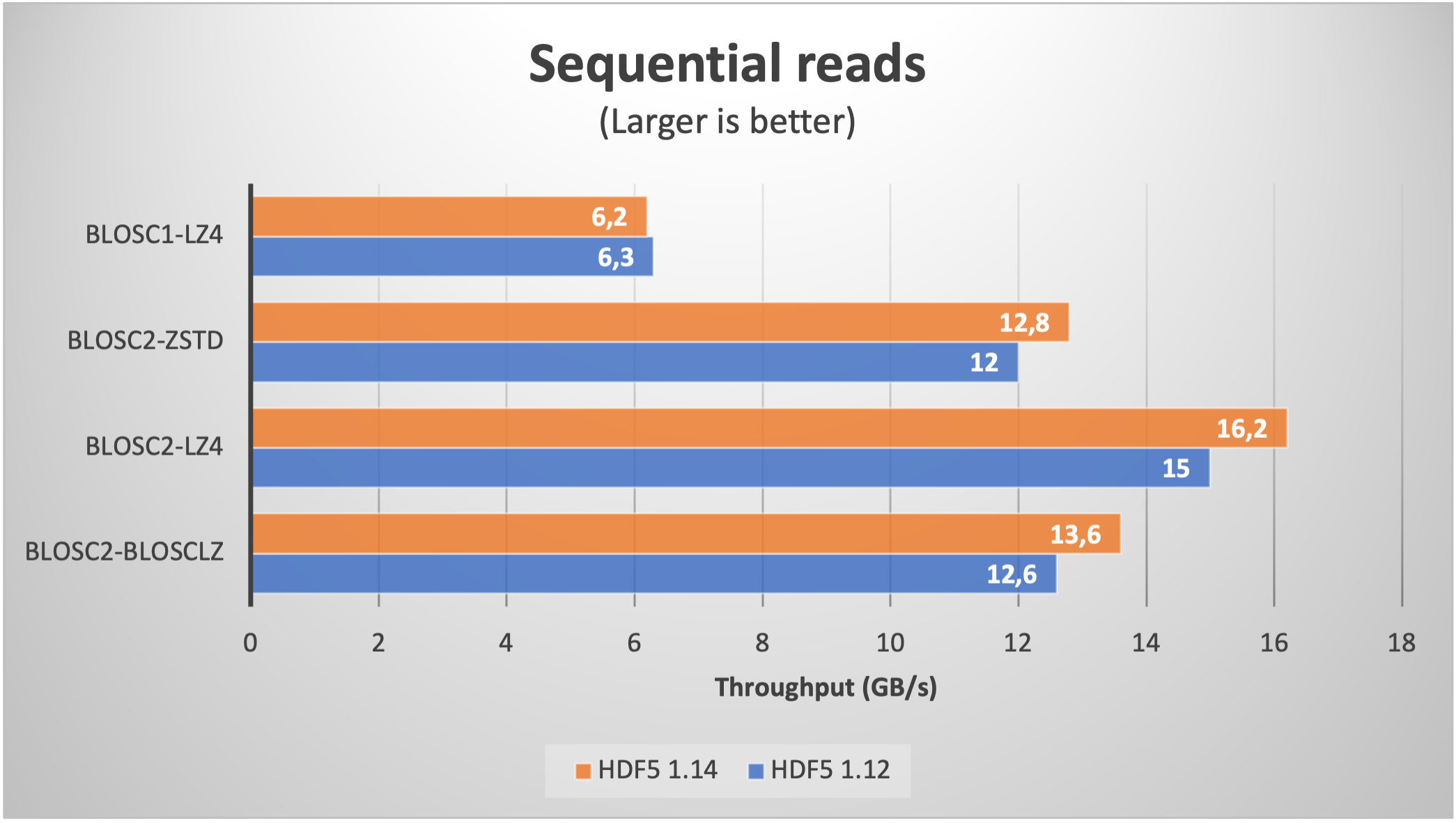
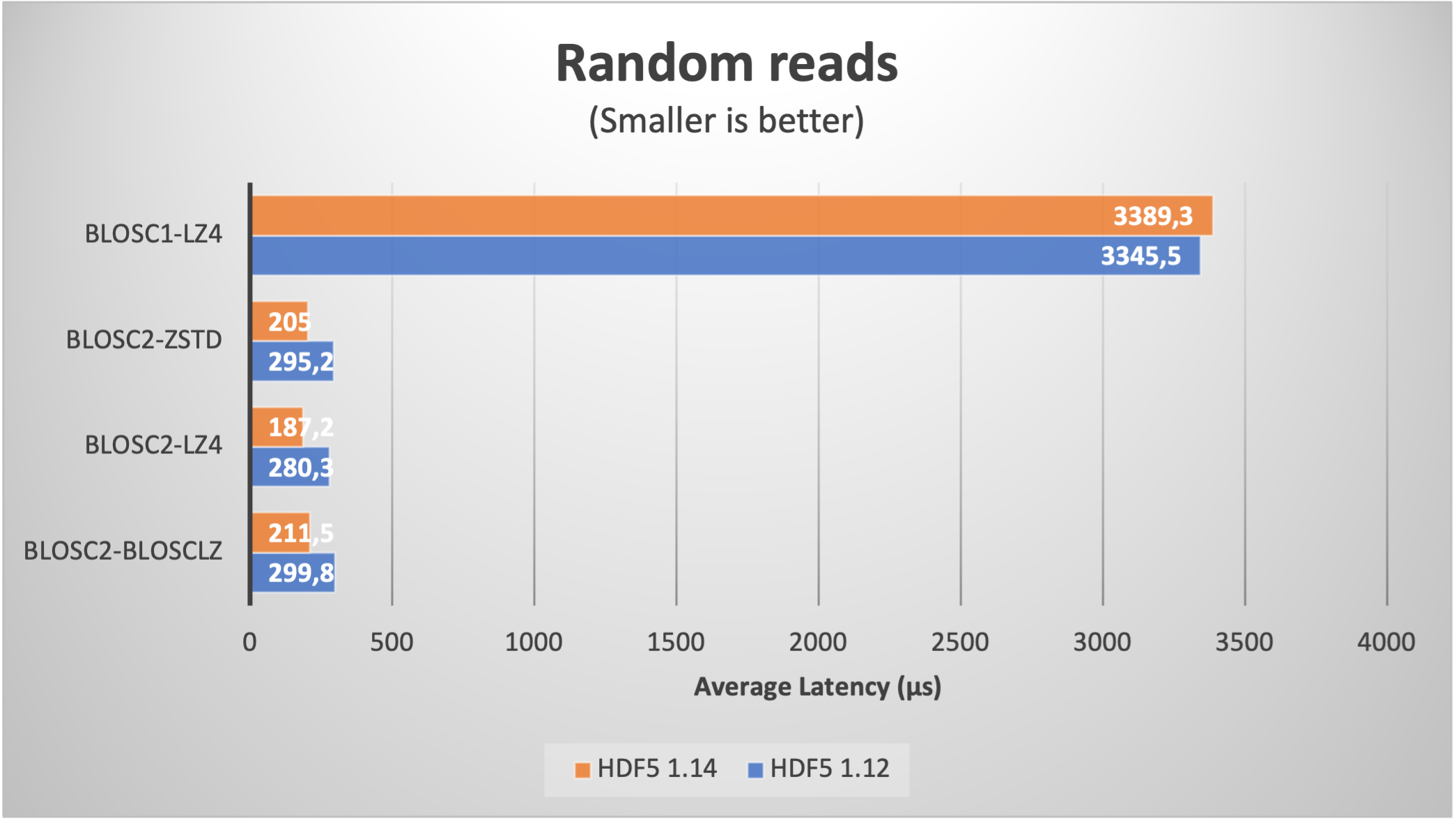
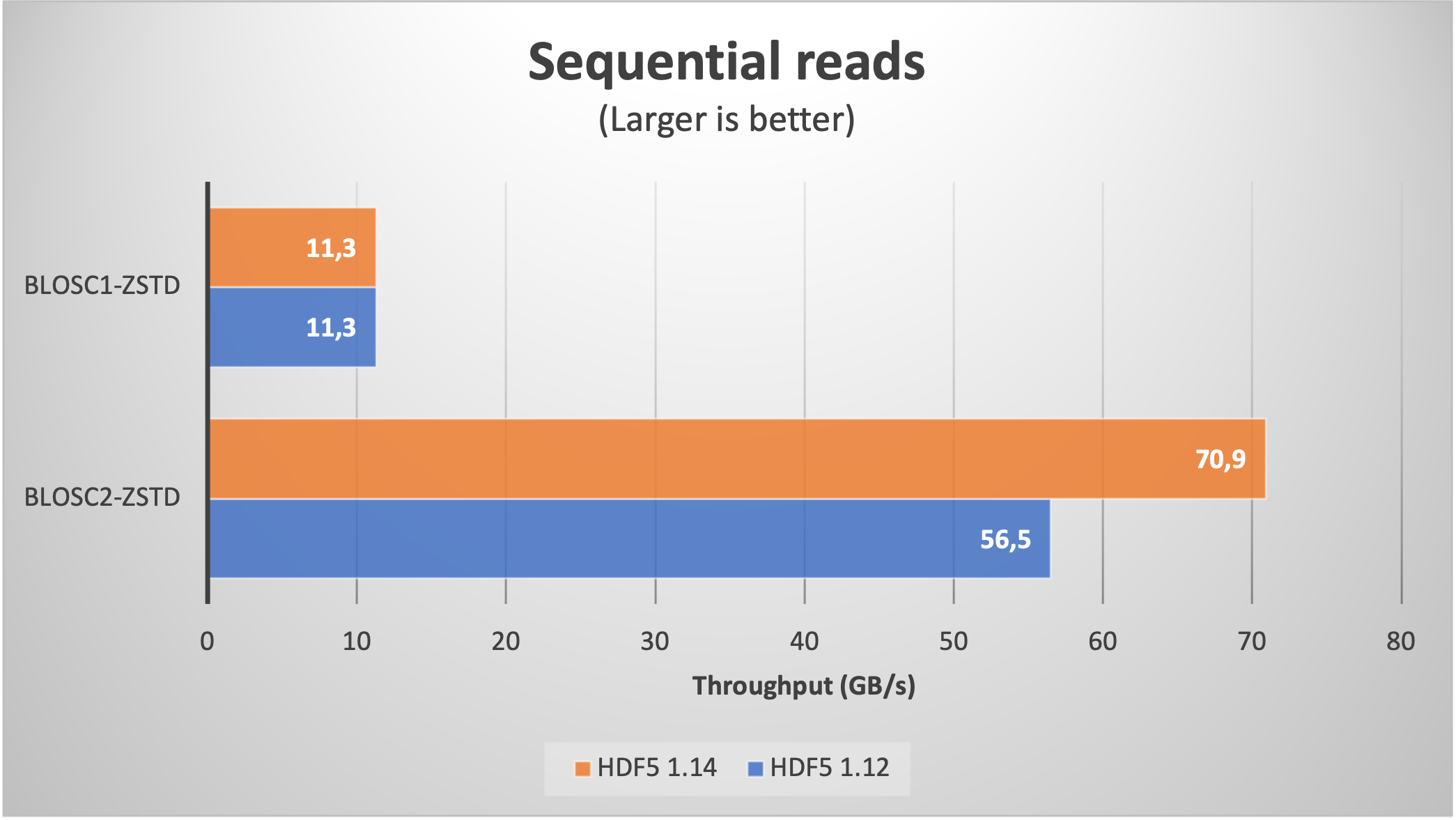
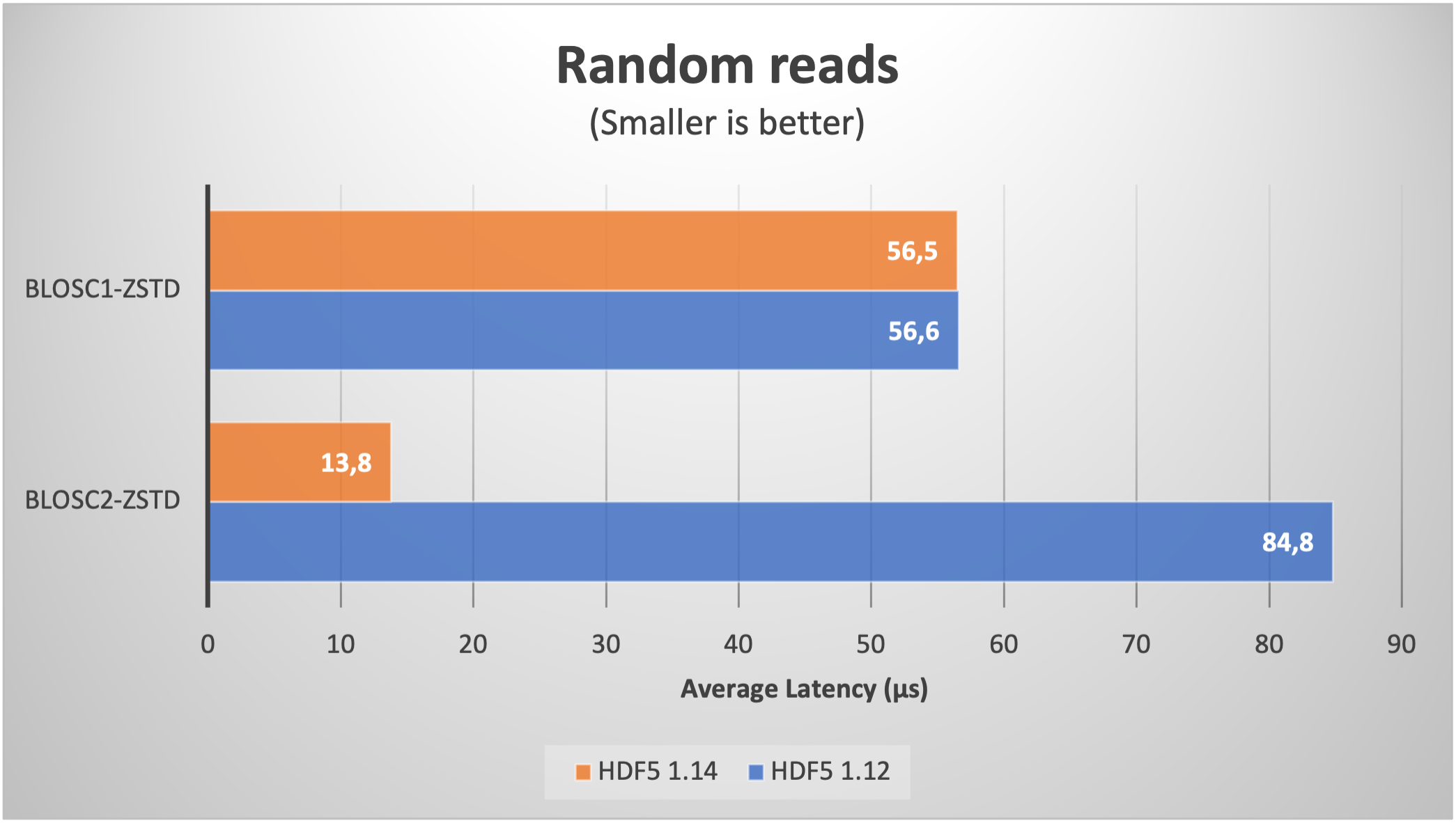
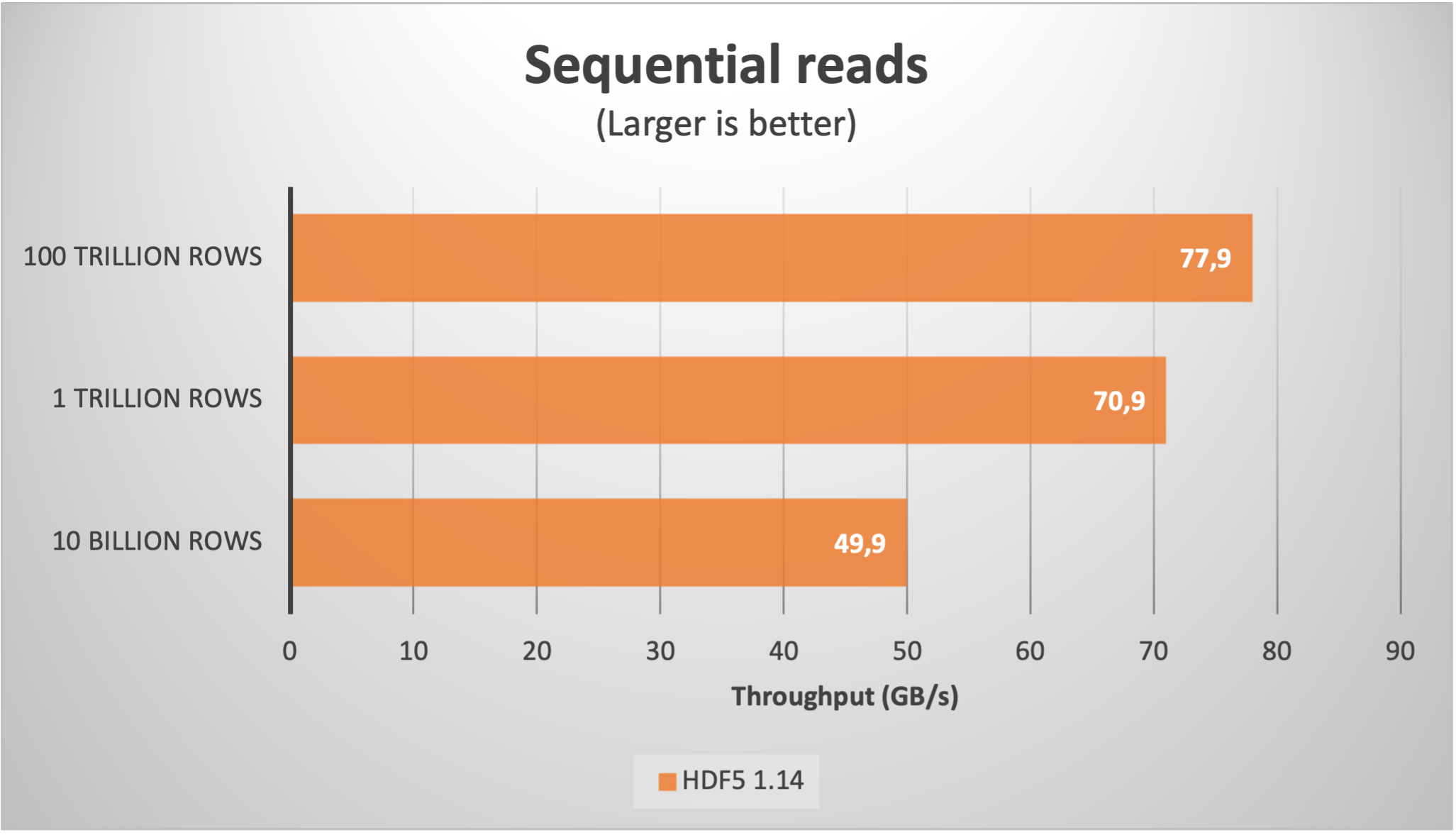
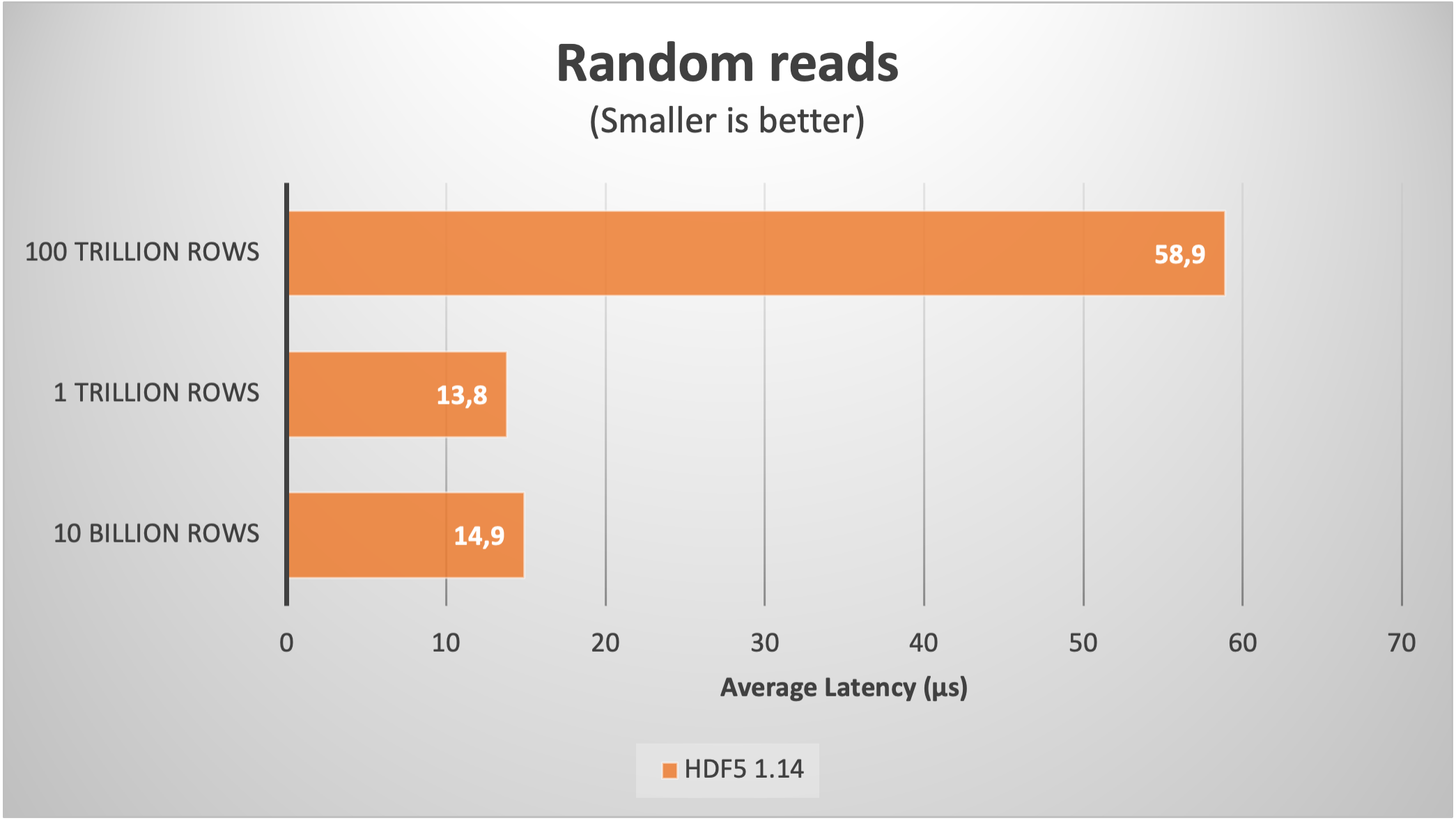
Using Blosc2 within HDF5
HDF5 is a widely used data format, and both major Python wrappers, h5py (via hdf5plugin) and PyTables, offer basic support for Blosc2. However, accessing the full capabilities of the Blosc2 compression pipeline is somewhat restricted because the current hdf5-blosc2 filter, available in PyTables (and used by hdf5plugin), is not yet equipped to transmit all the necessary parameters to the HDF5 data pipeline.
Thankfully, HDF5 includes support for the direct chunking mechanism, which enables the direct transmission of pre-compressed chunks to HDF5, bypassing its standard data pipeline. Since h5py also offers this functionality, it's entirely feasible to leverage all the advanced features of Blosc2, including lossy compression. Below are a couple of examples illustrating how this process operates:
Conclusion
Lossy compression is a powerful tool for optimizing storage space, reducing bandwidth usage, and improving overall efficiency in data handling. With Blosc2, developers have access to a robust and flexible compression library for both lossless and lossy compression modes.
With its advanced compression methodologies and adept memory management, Blosc2 empowers users to strike a harmonious balance between compression ratio, speed, and fidelity. This attribute renders it especially suitable for scenarios where resource limitations or performance considerations hold significant weight.
Finally, there are ongoing efforts towards integrating fidelity into our BTune AI tool. This enhancement will empower the tool to autonomously identify the most suitable codecs and filters, balancing compression level, precision, and fidelity according to user-defined preferences. Keep an eye out for updates!
Whether you're working with scientific data, multimedia content, or large-scale datasets, Blosc2 offers a comprehensive solution for efficient data compression and handling.
Special thanks to sponsors and developers
Gratitude goes out to our sponsors over the years, with special recognition to the LEAPS collaboration and NumFOCUS, whose support has been instrumental in advancing the lossy compression capabilities within Blosc2.
The Blosc2 project is the outcome of the work of many developers.