Using Proxies for Efficient Handling of Remote Multidimensional Data¶
When working with large datasets, a common problem is that they must be stored remotely, or on-disk, since they are too large to fit in memory. Doing so frees up memory for calculations with the data, but transfer times between the processor and the stored data can then cause bottlenecks. Blosc2 offers a way to manage this via proxies, and thus obtain the typical speedups associated with caching and in-memory storage of data, whilst still storing the dataset remotely/on-disk. This means we can mitigate the trade-off between storage space and execution time.
In this tutorial, we will look at how to access and cache data for calculation using the fetch and __getitem__ methods implemented in the Proxy class, the main Blosc2 proxy implementation. Through this comparison, we will gain a better understanding of how to optimize data access, as measured by the execution time of these retrieval operations. We will also measure the size of the local proxy file, to verify the efficiency of data management and storage. Get ready to dive into the
fascinating world of data caching!
[1]:
import asyncio
import os
import time
import blosc2
from blosc2 import ProxyNDSource
C2Array class¶
Before we look at proxies, it is first necessary to understand how to use Blosc2 to work with remote data, via the C2Array class. The class implements a (limited) version of the NDArray interface of which we have already seen a lot in previous tutorials. However, it is really a local pointer to a remote array (stored e.g. on a remote server). This means that we can refer to the data, access certain attribute information about it, download portions of the data and even define it in
computational expressions, without having to download the entire array into local memory or disk. This is particularly useful when working with large datasets that cannot fit into memory or would take far too long to transfer over the network.
However, one limitation of this approach is that every time one wants to download a slice of the dataset, the data is fetched over the network - even if the same slice has been downloaded before. This can lead to inefficiencies, especially when working with large datasets or when the same data is accessed multiple times. Proxies offer a solution to this, whilst still preserving the low storage requirements of the C2Array class.
Proxy Classes for Data Access¶
The `Proxy class <../../reference/proxy.rst>`__ in Blosc2 is a design pattern that acts as an intermediary between a (typically local) client and (typically remote or on-disk) real data containers, enabling more efficient access to the latter. Its primary objective is to provide a caching mechanism for effectively accessing data stored in remote/on-disk containers that utilize the ProxySource or ProxyNDSource interfaces, which serve as templates for defining custom proxy classes -
in themselves they cannot be used directly, as they are abstract classes.
We are going to define our own MyProxySource proxy class that will inherit from and implement the ProxyNDSource interface; it will be responsible for downloading and storing only the requested chunks, progressively filling the cache as the user accesses the data.
[2]:
def get_file_size(filepath):
"""Returns the file size in megabytes."""
return os.path.getsize(filepath) / (1024 * 1024)
class MyProxySource(ProxyNDSource):
def __init__(self, data):
self.data = data
print(f"Data shape: {self.shape}, chunks: {self.chunks}, dtype: {self.dtype}")
@property
def shape(self):
return self.data.shape
@property
def chunks(self):
return self.data.chunks
@property
def blocks(self):
return self.data.blocks
@property
def dtype(self):
return self.data.dtype
# This method must be present
def get_chunk(self, nchunk):
return self.data.get_chunk(nchunk)
# This method is optional
async def aget_chunk(self, nchunk):
await asyncio.sleep(0.1) # simulate an asynchronous operation
return self.data.get_chunk(nchunk)
Next, we will establish a connection to a multidimensional array stored remotely on a Cat2Cloud demo server (https://cat2.cloud/demo). The remote_array variable will represent this dataset on the server, via a C2Array, enabling us to access the information without the need to load all the data into local memory at once.
[3]:
urlbase = "https://cat2.cloud/demo"
path = "@public/examples/lung-jpeg2000_10x.b2nd"
remote_array = blosc2.C2Array(path, urlbase=urlbase)
Although it is not as useful, note that a MyProxySource instance could also be constructed with an NDArray object stored on-disk, so that one can cache parts of the array in-memory for quicker access. In either case, the data of the C2Array/NDArray is linked by the MyProxySource instance to a local Proxy instance (instantiated using the source) acting as an in-memory cache for the data.
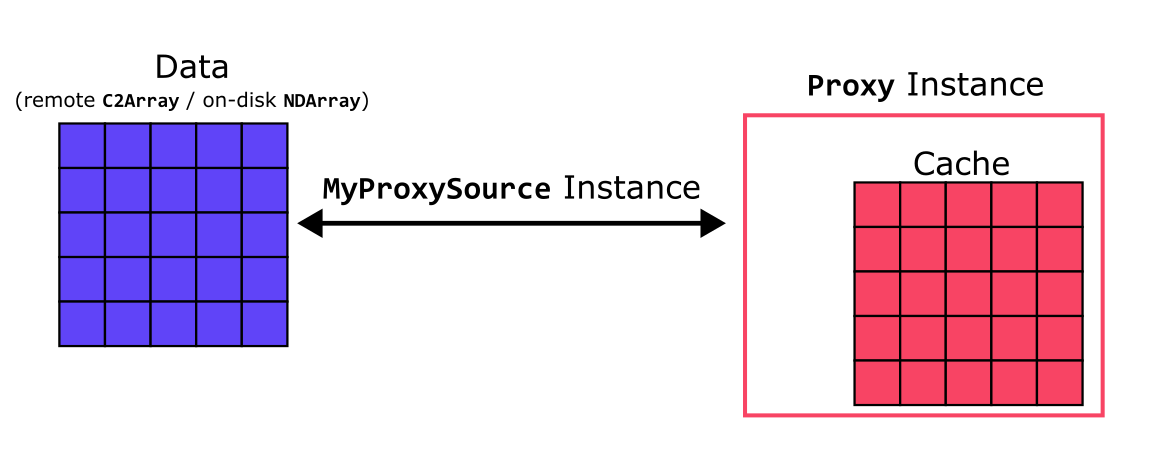
[4]:
# Define a local file path to save the proxy container
local_path = "local_proxy_container.b2nd"
source = MyProxySource(remote_array)
proxy = blosc2.Proxy(source, urlpath=local_path, mode="w")
print(f"Proxy of type {type(proxy)} has shape {proxy.shape}, chunks {proxy.chunks} and dtype {proxy.dtype}")
initial_size = get_file_size(local_path)
print(f"Initial local file size: {os.path.getsize(local_path)} bytes")
Data shape: (10, 1248, 2689), chunks: (1, 1248, 2689), dtype: uint16
Proxy of type <class 'blosc2.proxy.Proxy'> has shape (10, 1248, 2689), chunks (1, 1248, 2689) and dtype uint16
Initial local file size: 321 bytes
As can be seen, the local proxy container occupies a few hundred bytes, which is significantly smaller than the remote dataset (around 64 MB, 6.4 MB compressed). This is because the local container only contains metadata about the remote dataset, such as its shape and data type, but not the actual data. The proxy will download the data from the remote source as needed, storing it in the local container for future access.
Retrieving data with a Proxy¶
The Proxy class implements two methods to retrieve data: fetch and __getitem__. Similar to the NDArray methods slice (returns NDArray) and __getitem__ (returns NumPy array) fetch returns an NDArray and __getitem__ a NumPy array. However, there are more differences, which we’ll now detail.
The fetch method¶
fetch is designed to return the full local proxy (with shape the same as the source data), which serves as a cache for the requested data. The cache is initialized with zeros in all entries, before the first fetch call; when fetch is called with a specific slice, the required chunks are downloaded from the remote source and used to populate the relevant entries in the local proxy container; the remaining entries remain uninitialized with zeros. If fetch is called again with a
different slice, only the new chunks necessary to fil out the new slice are downloaded to fill the relevant entries of the cache. If the same slice is requested again, the data is already present in the local proxy cache, so the cache is returned immediately with no download occurring.
In this way, fetch downloads only the specific data that is required, which reduces the amount of data stored locally and optimizes the use of resources. This method is particularly useful when working with large datasets, as it allows for the efficient handling of multidimensional data.
[5]:
# Fetch a slice of the data from the proxy
t0 = time.time()
slice_data = proxy.fetch(slice(0, 2))
t1 = time.time() - t0
print(f"Time to fetch: {t1:.2f} s")
print(f"slice_data is of type {type(slice_data)} and shape {slice_data.shape}.")
print(f"File size after fetch (2 chunks): {get_file_size(local_path):.2f} MB")
print(slice_data[1:3, 1:3])
Time to fetch: 0.92 s
slice_data is of type <class 'blosc2.ndarray.NDArray'> and shape (10, 1248, 2689).
File size after fetch (2 chunks): 1.28 MB
[[[15712 13933 18298 ... 21183 22486 20541]
[18597 21261 23925 ... 22861 21008 19155]]
[[ 0 0 0 ... 0 0 0]
[ 0 0 0 ... 0 0 0]]]
Above, using the fetch function with a slice involves downloading data from a chunk that had not been previously requested, increasing the local file size as new data is stored. fetch returns the local proxy cache as an NDArray instance into the slice_data variable.
In the previous result, only the 2 chunks necessary (to understand why two chunks are necessary, look at the chunk shape) to fill the desired slice slice(0, 2) have been downloaded and initialized, which is reflected in the array with visible numerical values, as seen in the section [[15712 13933 18298 ... 21183 22486 20541], [18597 21261 23925 ... 22861 21008 19155]]. These represent data that are ready to be processed.
On the other hand, the lower part of the array, [[0 0 0 ... 0 0 0], [0 0 0 ... 0 0 0]], shows an uninitialized section of the proxy (normally filled with zeros). This indicates that those chunks have not yet been downloaded or processed. The fetch function could eventually fill these chunks with data when requested, replacing the zeros (which indicate uninitialized data) with the corresponding values:
[6]:
# Fetch a slice of the data from the proxy
t0 = time.time()
slice_data2 = proxy.fetch((slice(2, 3), slice(6, 7)))
t1 = time.time() - t0
print(f"Time to fetch: {t1:.2f} s")
print(f"File size after fetch (1 chunk): {get_file_size(local_path):.2f} MB")
print(slice_data[1:3, 1:3])
Time to fetch: 0.44 s
File size after fetch (1 chunk): 1.92 MB
[[[15712 13933 18298 ... 21183 22486 20541]
[18597 21261 23925 ... 22861 21008 19155]]
[[16165 14955 19889 ... 21203 22518 20564]
[18610 21264 23919 ... 20509 19364 18219]]]
Now the fetch function has downloaded another additional chunk, which is reflected in the local file size. We can also see that now the slice [1:3, 1:3] has been initialized with data, while the rest of the proxy array will remain uninitialized.
The __getitem__ method¶
The __getitem__ function in the Proxy class is similar to fetch in that it allows for the retrieval of specific data from the remote container. However, __getitem__ returns a NumPy array which only contains the explicitly requested data (and not the whole proxy with initialized and uninitialized entries).
[7]:
# Using __getitem__ to get a slice of the data
t0 = time.time()
result = proxy[5:7, 1:3]
t1 = time.time() - t0
print(f"Proxy __getitem__ time: {t1:.3f} s")
print(f"result is of type {type(result)} and shape {result.shape}.")
print(f"File size after __getitem__ (2 chunks): {get_file_size(local_path):.2f} MB")
Proxy __getitem__ time: 0.891 s
result is of type <class 'numpy.ndarray'> and shape (2, 2, 2689).
File size after __getitem__ (2 chunks): 3.20 MB
However, behind the scenes fetch is called, since the relevant chunks have not been initialized, and these are then downloaded to the cache - hence the size of the local file has increased. The __getitem__ function then retrieves and decompresses the data in the chunks stored in the proxy container, and returns the slice into the result array, which is now available for processing.
Differences between fetch and __getitem__¶
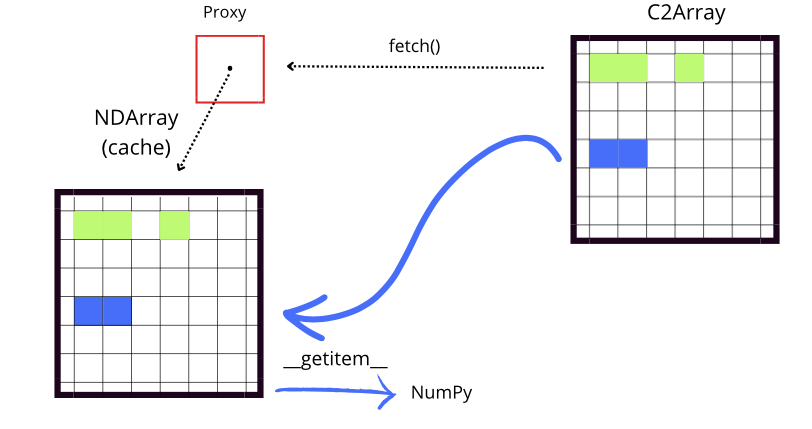
Although fetch and __getitem__ have distinct functions, they work together to facilitate efficient access to data. fetch manages the loading of data into the local cache by checking if the necessary chunks are available. If they are not, it downloads them from the remote source i to the proxy cache for future access.
On the other hand, __getitem__ handles the indexing and retrieval of data through a NumPy array, allowing access to specific subsets. Before accessing the data, __getitem__ calls fetch to ensure that the necessary chunks are in the cache. If the data is not present in the cache, fetch takes care of downloading it from its original location (for example, from disk or an external source). This ensures that when __getitem__ performs the indexing operation, it has immediate
access to the data without interruptions.
An important detail is that, while both fetch and __getitem__ ensure the necessary data is available, they may download more information than required because they download entire chunks (and not just the required slice). However, this can be advantageous for two reasons. Firstly, often one wants to access multiple slices of large remote arrays within a script, and thus slices may overlap with already-downloaded chunks from a previous fetch; by fetching the whole chunk in the first
slice, one already has the data locally for future slice commands, thus implementing an efficient data prefetcher. Secondly, by sending the whole (compressed) chunk, the data is always compressed during the complete workflow (file transfer and storage), which reduces storage space, file transfer time, and processing overheads.
[8]:
# clean up
blosc2.remove_urlpath("local_proxy_container.b2nd")
About the remote dataset¶
The remote dataset is available online. You may want to explore the data values by clicking on the Data tab; this dataset is actually a tomography of a lung, which you can visualize by clicking on the Tomography tab. Finally, by clicking on the Download button, the file can be downloaded locally in case you want to experiment more with the data.
As we have seen, every time that we downloaded a chunk, the size of the local file increased by a fixed amount (around 0.64 MB). This is because the chunks (whose uncompressed data occupies around 6.4 MB) are compressed with the Codec.GROK codec, which has been configured to reduce the size of the data by a constant factor of 10. This means that the compressed data occupies only one-tenth of the space that it would occupy without compression. This reduction in data size optimizes both
storage and transfer, as data is always handled in a compressed state when downloading or storing images, accelerating the transfer process (by a factor of 10).
Conclusion¶
This tutorial has highlighted how the Proxy class in Blosc2, combined with the fetch and __getitem__ methods, optimizes access to multidimensional data, even when stored remotely (accessible via a C2Array). The intelligent use of a workflow which links remote/on-disk data (C2Array/NDArray) to a local Proxy cache (via a ProxyNDSource instance) enables one to handle large volumes of information without maxing out storage capacity, whilst still benefitting from the
performance gains of caching and in-memory calculation.