TreeStore: Endowing Your Data With Hierarchical Structure
When working with large and complex datasets, having a way to organize your data efficiently is crucial. blosc2.TreeStore is a powerful feature in the blosc2 library that allows you to store and manage your compressed arrays in a hierarchical, tree-like structure, much like a filesystem. This container, typically saved with a .b2z extension, can hold not only blosc2.NDArray or blosc2.SChunk objects but also metadata, making it a versatile tool for data organization.
What is a TreeStore?
A TreeStore lets you arrange your data into groups (like directories) and datasets (like files). Each dataset is a blosc2.NDArray or blosc2.SChunk instance, benefiting from Blosc2's high-performance compression. This structure is ideal for scenarios where data has a natural hierarchy, such as in scientific experiments, simulations, or any project with multiple related datasets.
Basic Usage: Creating and Populating a TreeStore
Creating a TreeStore is straightforward. You can use a with statement to ensure the store is properly managed. Inside the with block, you can create groups and datasets using a path-like syntax.
import blosc2 import numpy as np # Create a new TreeStore with blosc2.TreeStore("my_experiment.b2z", mode="w") as ts: # You can store numpy arrays, which are converted to blosc2.NDArray ts["/dataset0"] = np.arange(100) # Create a group with a dataset that can be a blosc2 NDArray ts["/group1/dataset1"] = blosc2.zeros((10,)) # You can also store blosc2 arrays directly (vlmeta included) ext = blosc2.linspace(0, 1, 10_000, dtype=np.float32) ext.vlmeta["desc"] = "dataset2 metadata" ts["/group1/dataset2"] = ext
In this example, we created a TreeStore in a file named my_experiment.b2z.
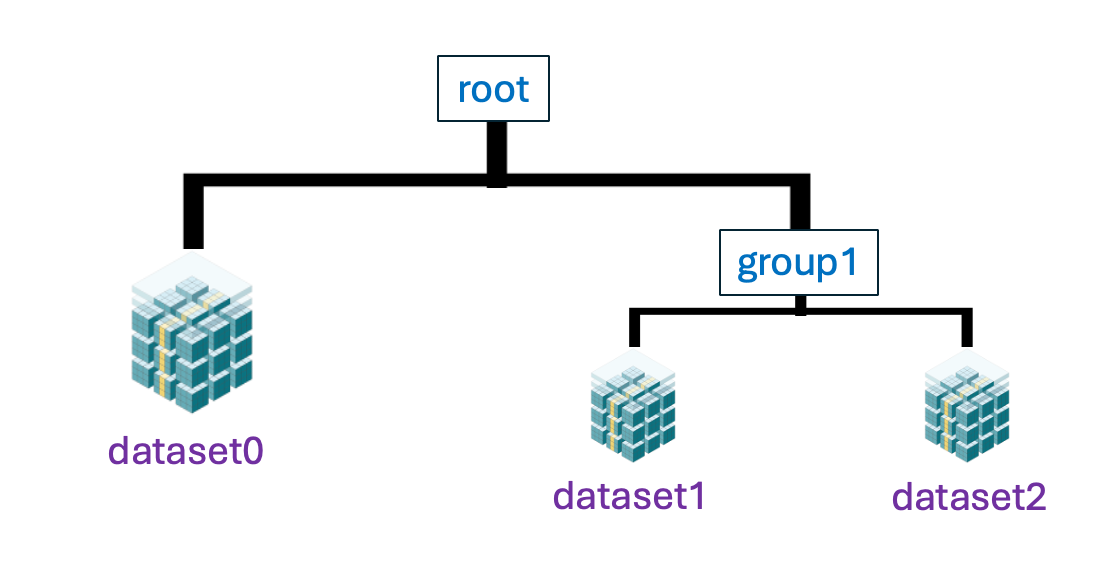
It contains two groups, root and group1, each holding datasets.
Reading from a TreeStore
To access the data, you open the TreeStore in read mode ('r') and use the same path-like keys to retrieve your arrays.
# Open the TreeStore in read-only mode ('r') with blosc2.TreeStore("my_experiment.b2z", mode="r") as ts: # Access a dataset dataset1 = ts["/group1/dataset1"] print("Dataset 1:", dataset1[:]) # Use [:] to decompress and get a NumPy array # Access the external array that has been stored internally dataset2 = ts["/group1/dataset2"] print("Dataset 2", dataset2[:]) print("Dataset 2 metadata:", dataset2.vlmeta[:]) # List all paths in the store print("Paths in TreeStore:", list(ts))
Advanced Usage: Metadata and Subtrees
TreeStore becomes even more powerful when you use metadata and interact with subtrees (groups).
Storing Metadata with vlmeta
You can attach variable-length metadata (vlmeta) to any group or to the root of the tree. This is useful for storing information like author names, dates, or experiment parameters. vlmeta is essentially a dictionary where you can store your metadata.
# Appending metadata to the TreeStore with blosc2.TreeStore("my_experiment.b2z", mode="a") as ts: # 'a' for append/modify # Add metadata to the root ts.vlmeta["author"] = "The Blosc Team" ts.vlmeta["date"] = "2025-08-17" # Add metadata to a group ts["/group1"].vlmeta["description"] = "Data from the first run" # Reading metadata with blosc2.TreeStore("my_experiment.b2z", mode="r") as ts: print("Root metadata:", ts.vlmeta[:]) print("Group 1 metadata:", ts["/group1"].vlmeta[:])
Working with Subtrees (Groups)
A group object can be retrieved from the TreeStore and treated as a smaller, independent TreeStore. This capability is useful for better organizing your data access code.
with blosc2.TreeStore("my_experiment.b2z", mode="r") as ts: # Get the group as a subtree group1 = ts["/group1"] # Now you can access datasets relative to this group dataset2 = group1["dataset2"] print("Dataset 2 from group object:", dataset2[:]) # You can also list contents relative to the group print("Contents of group1:", list(group1))
Iterating Through a TreeStore
You can easily iterate through all the nodes in a TreeStore to inspect its contents.
with blosc2.TreeStore("my_experiment.b2z", mode="r") as ts: for path, node in ts.items(): if isinstance(node, blosc2.NDArray): print(f"Found dataset at '{path}' with shape {node.shape}") else: # It's a group print(f"Found group at '{path}' with metadata: {node.vlmeta[:]}")
Found dataset at '/group1/dataset2' with shape (10000,) Found group at '/group1' with metadata: {'description': 'Data from the first run'} Found dataset at '/group1/dataset1' with shape (10,) Found dataset at '/dataset0' with shape (100,)
That's it for this introduction to blosc2.TreeStore! You now know how to create, read, and manipulate a hierarchical data structure that can hold compressed datasets and metadata. You can find the source code for this example in the blosc2 repository.
Some Benchmarks
TreeStore is based on powerful abstractions from the blosc2 library, so it is very fast. Here are some benchmarks comparing TreeStore to other data storage formats, like HDF5 and Zarr. We have used two different configurations: one with small arrays, where sizes follow a normal distribution centered at 10 MB each, and the other with larger arrays, where sizes follow a normal distribution centered at 1 GB each. We have compared the performance of TreeStore against HDF5 and Zarr for both small and large arrays, measuring the time taken to create and read datasets. For comparing apples with apples, we have used the same compression codec (zstd) and filter (shuffle) for all three formats.
For assessing different platforms, we have used a desktop with an Intel i9-13900K CPU and 32 GB of RAM, running Ubuntu 25.04, and also a Mac mini with an Apple M4 Pro processor and 24 GB of RAM. The benchmarks were run using this script.
Results for the Intel i9-13900K desktop
100 small arrays (around 10 MB each) scenario:
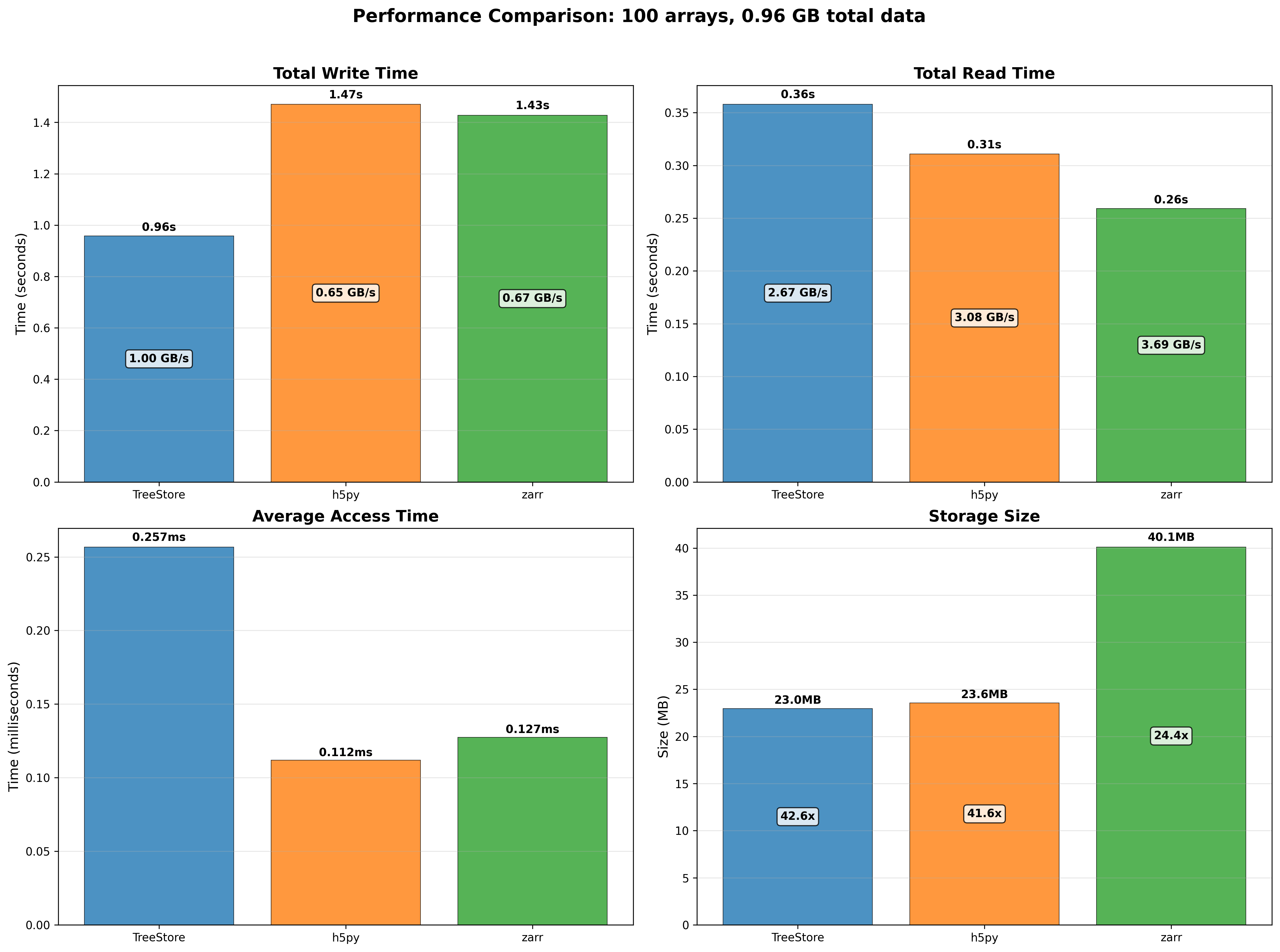
For the small arrays scenario, we can see that TreeStore is the fastest to create datasets (due to use of multi-threading), but it is slower than HDF5 and Zarr when reading datasets. The reason for this is two-fold: first, TreeStore is designed to work using multi-threading, so it must setup the necessary threads at the beginning of the read operation, which takes some time; second, TreeStore is using NDArray objects internally, which are using a double partitioning scheme (chunks and blocks) to store the data, which adds some overhead when reading small slices of data. Regarding the space used, TreeStore is the most efficient, very close to HDF5, and significantly more efficient than Zarr.
100 large arrays (around 1 GB each) scenario:
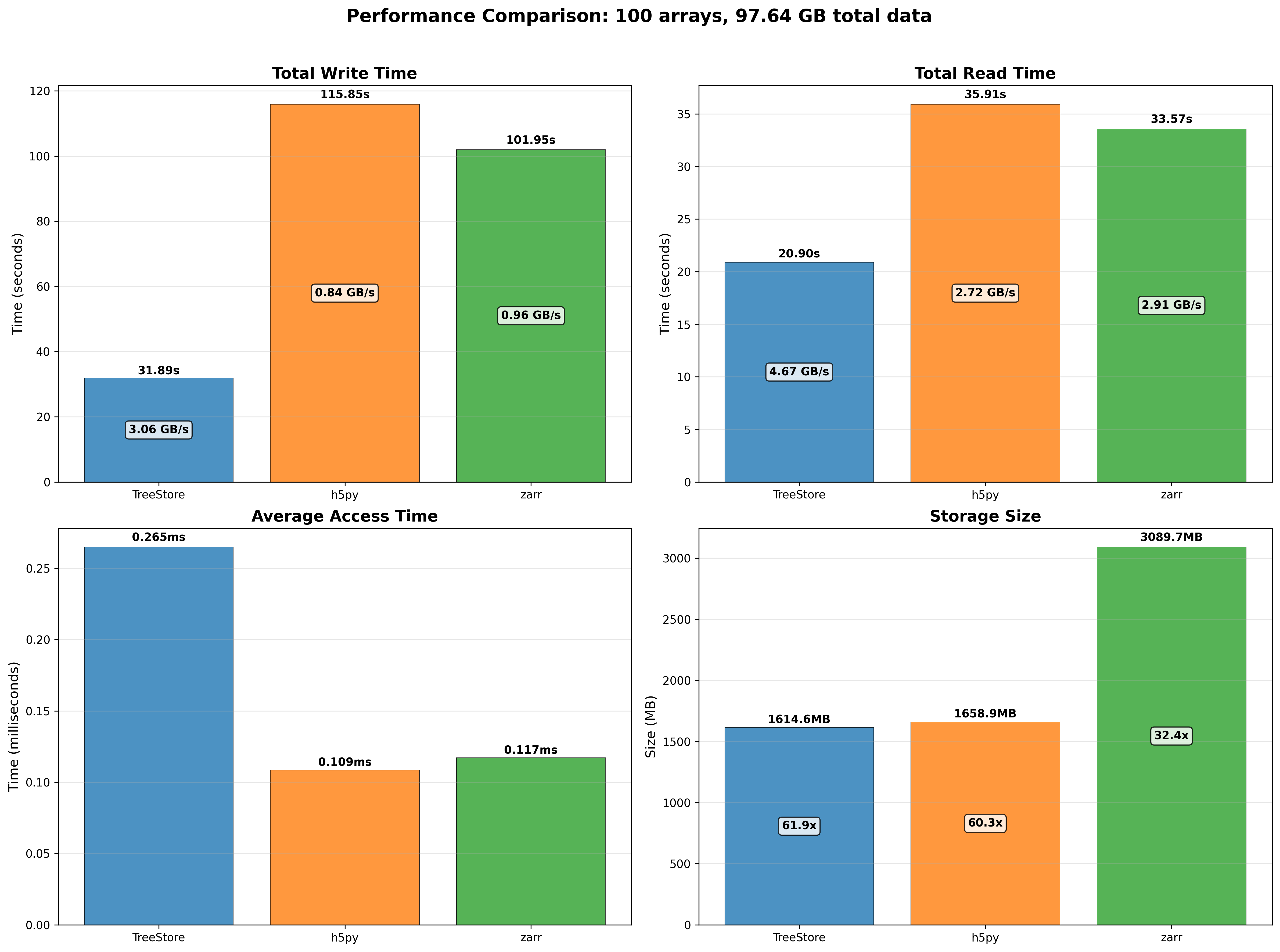
When handling larger arrays, TreeStore maintains its lead in creation and full-read performance. Although HDF5 and Zarr offer faster access to small data slices, TreeStore compensates by being the most storage-efficient format, followed by HDF5, with Zarr being the most space-intensive.
Results for the Apple M4 Pro Mac mini
100 small arrays (around 10 MB each) scenario:
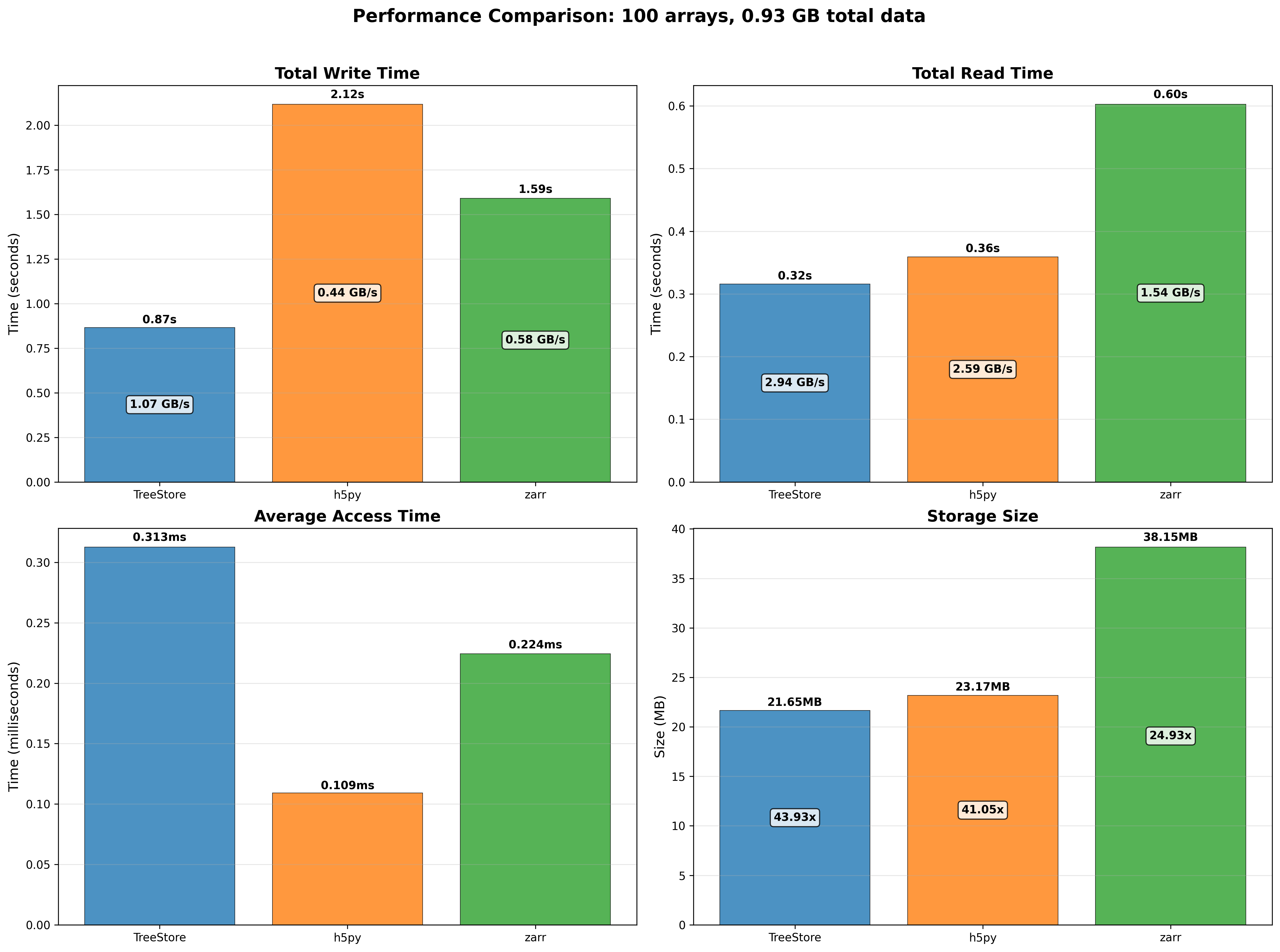
100 large arrays (around 1 GB each) scenario:
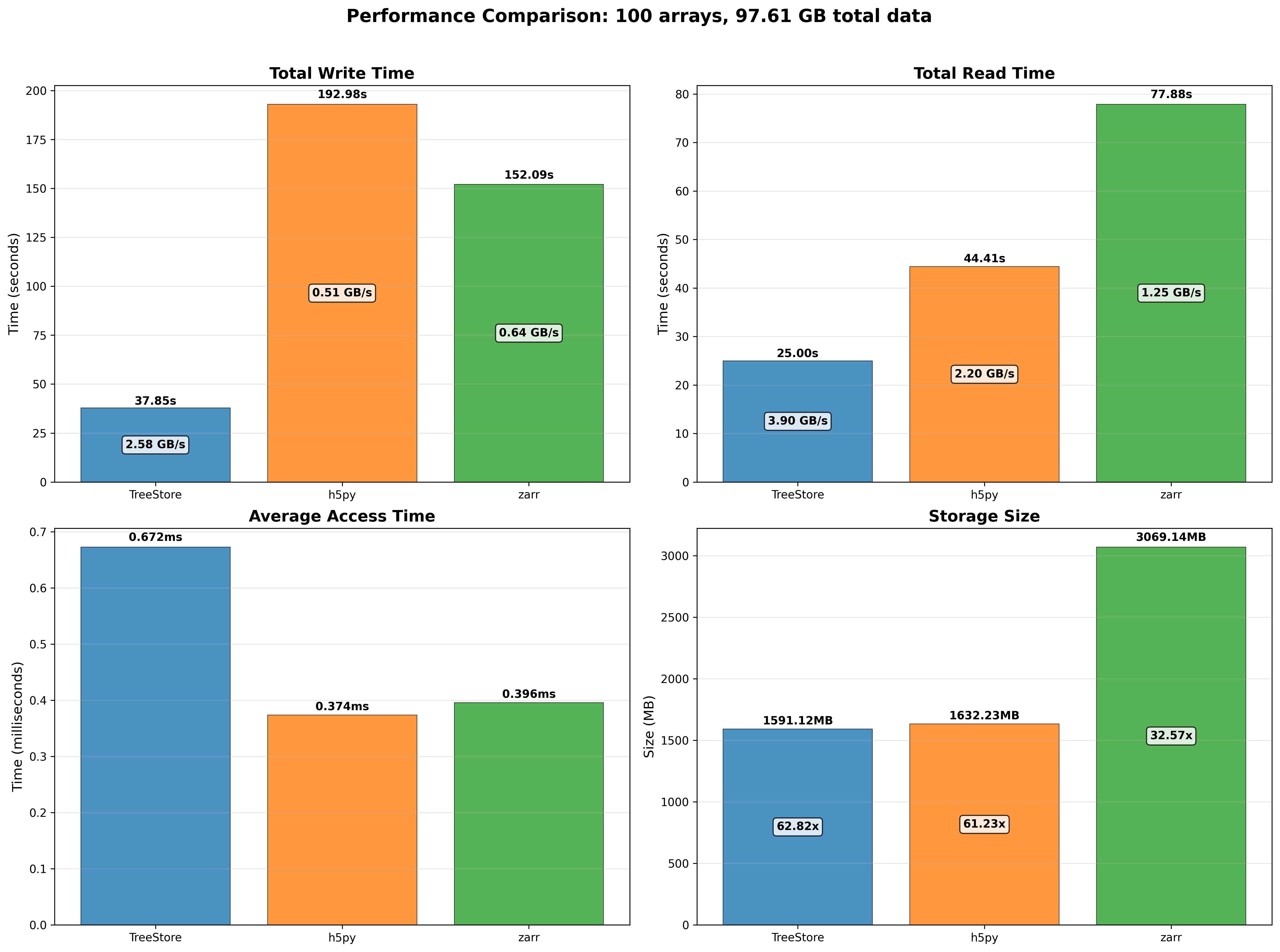
Consistent with the previous results, TreeStore is the most space-efficient format and the fastest for creating and reading datasets, particularly for larger arrays. Its performance is slower than HDF5 and Zarr only when reading small data slices (access time). This can be improved by reducing the number of threads from the default of eight, which lessens the thread setup overhead. For more details on this, see these slides comparing 8-thread vs 1-thread performance.
Notably, the Apple M4 Pro processor shows competitive performance against the Intel i9-13900K CPU, a high-end desktop processor that consumes up to 8x more power. This result underscores the efficiency of the ARM architecture in general and Apple silicon in particular.
Conclusion
In summary, blosc2.TreeStore offers a straightforward yet potent solution for hierarchically organizing compressed datasets. By merging the high-performance compression of blosc2.NDArray and blosc2.SChunk with a flexible, filesystem-like structure and metadata support, it stands out as an excellent choice for managing complex data projects.
As TreeStore is currently in beta, we welcome feedback and suggestions for its improvement. For further details, please consult the official documentation for blosc2.TreeStore.
Comments
Comments powered by Disqus